Abstract
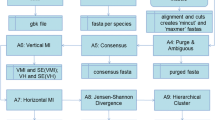
The paper introduces a novel probability descriptor for genome sequence comparison, employing a generalized form of Jensen-Shannon divergence. This divergence metric stems from a one-parameter family, comprising fractions up to a maximum value of half. Utilizing this metric as a distance measure, a distance matrix is computed for the new probability descriptor, shaping Phylogenetic trees via the neighbor-joining method. Initial exploration involves setting the parameter at half for various species. Assessing the impact of parameter variation, trees drawn at different parameter values (half, one-fourth, one-eighth). However, measurement scales decrease with parameter value increments, with higher similarity accuracy corresponding to lower scale values. Ultimately, the highest accuracy aligns with the maximum parameter value of half. Comparative analyses against previous methods, evaluating via Symmetric Distance (SD) values and rationalized perception, consistently favor the present approach's results. Notably, outcomes at the maximum parameter value exhibit the most accuracy, validating the method's efficacy against earlier approaches.






Similar content being viewed by others
Data Availability
No datasets were generated or analysed during the current study.
References
Phillips A, Janies D, Wheeler W (2000) Multiple sequence alignment in phylogenetic analysis. Mol Phylogenet Evol 16(3):317–330. https://doi.org/10.1006/mpev.2000.0785
Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22(22):4673–4680. https://doi.org/10.1093/nar/22.22.4673
Katoh K et al (2002) MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. https://doi.org/10.1093/nar/gkf436
Vinga S, Almeida J (2003) Alignment-free sequence comparison—A review. Bioinformatics 19(4):513–523. https://doi.org/10.1093/bioinformatics/btg005
Domazet-Lošo M, Haubold B (2011) Alignment-free detection of local similarity among viral and bacterial genomes. Bioinformatics 27(11):1466–1472. https://doi.org/10.1093/bioinformatics/btr176
Gates MA (1986) A simple way to look at DNA. J Theor Biol 119(3):319–328. https://doi.org/10.1016/s0022-5193(86)80144-8
Nandy A (1994) A new graphical representation and analysis of DNA sequence structure: I. Methodology and application to globin genes. Curr Sci 66:309–314
Leong PM, Morgenthaler S (1995) Random walk and gap plots of DNA sequences. Bioinformatics 11(5):503–507. https://doi.org/10.1093/bioinformatics/11.5.503
Guo X, Randic M, Basak SC (2001) A novel 2-D graphical representation of DNA sequences of low degeneracy. Chem Phys Lett 350(1–2):106–112. https://doi.org/10.1016/S0009-2614(01)01246-5
Yau SS et al (2003) DNA sequence representation without degeneracy. Nucleic Acids Res 31(12):3078–3080. https://doi.org/10.1093/nar/gkg432
Liao Bo (2005) A 2D graphical representation of DNA sequence. Chem Phys Lett 401(1–3):196–199. https://doi.org/10.1016/j.cplett.2004.11.059
Liao Bo, Tan M, Ding K (2005) Application of 2-D graphical representation of DNA sequence. Chem Phys Lett 414(4–6):296–300. https://doi.org/10.1016/J.CPLETT.2005.08.079
Song J, Tang H (2005) A new 2-D graphical representation of DNA sequences and their numerical characterization. J Biochem Biophys Methods 63(3):228–239. https://doi.org/10.1016/j.jbbm.2005.04.004
Randić M et al (2003) Novel 2-D graphical representation of DNA sequences and their numerical characterization. Chem Phys Lett 368(1–2):1–6. https://doi.org/10.1016/S0009-2614(02)01784-0
Randić M et al (2003) Analysis of similarity/dissimilarity of DNA sequences based on novel 2-D graphical representation. Chem Phys Lett 371(1–2):202–207. https://doi.org/10.1016/S0009-2614(03)00244-6
Yao Y-H, Liao Bo, Wang T-M (2005) A 2D graphical representation of RNA secondary structures and the analysis of similarity/dissimilarity based on it. J Mol Struct (Thoechem) 755(1–3):131–136. https://doi.org/10.1016/j.theochem.2005.08.009
Randić M et al (2000) On 3-D graphical representation of DNA primary sequences and their numerical characterization. J Chem Inf Comput Sci 40(5):1235–1244. https://doi.org/10.1021/ci000034q
Nandy A, Nandy P (1995) Graphical analysis of DNA sequence structure: II. Relative abundances of nucleotides in DNAs, gene evolution and duplication. Curr Sci 68:75–85
Yao Y-H, Nan X-Y, Wang T-M (2006) A new 2D graphical representation—Classification curve and the analysis of similarity/dissimilarity of DNA sequences. J Mol Struct (Thoechem) 764(1–3):101–108. https://doi.org/10.1016/j.theochem.2006.02.007
Das S, Pal J, Bhattacharya DK (2015) Geometrical method of exhibiting similarity/dissimilarity under new 3D classification curves and establishing significance difference of different parameters of estimation. Intl J Adv Res Comp Sci SoftwEngg 5:279–287
Randić M et al (2001) On characterization of proteomics maps and chemically induced changes in proteomes using matrix invariants: application to peroxisome proliferators. Med Chem Res 10(7–8):456–479
Qi Z-H, Fan T-R (2007) PN-curve: A 3D graphical representation of DNA sequences and their numerical characterization. Chem Phys Lett 442(4–6):434–440. https://doi.org/10.1016/j.cplett.2007.06.029
Akhtar M, Epps J, Ambikairajah E (2008) Signal processing in sequence analysis: advances in eukaryotic gene prediction. IEEE J Selected Topics Signal Process 2(3):310–321. https://doi.org/10.1109/JSTSP.2008.923854
Chakravarthy N et al (2004) Autoregressive modeling and feature analysis of DNA sequences. EURASIP J Adv Signal Process 2004(1):1–16. https://doi.org/10.1155/S111086570430925X
Chi R, Ding K (2005) Novel 4D numerical representation of DNA sequences. Chem Phys Lett 407(1–3):63–67. https://doi.org/10.1016/j.cplett.2005.03.056
Nieto JJ, Torres A, Vázquez-Trasande MM (2003) A metric space to study differences between polynucleotides. Appl Math Lett 16(8):1289–1294. https://doi.org/10.1016/S0893-9659(03)90131-5
Nieto JJ et al (2006) Fuzzy polynucleotide spaces and metrics. Bull Math Biol 68(3):703–725. https://doi.org/10.1007/s11538-005-9020-5
Torres A, Nieto JJ (2003) The fuzzy polynucleotide space: basic properties. Bioinformatics 19(5):587–592. https://doi.org/10.1093/bioinformatics/btg032
Sadegh-Zadeh K (2000) Fuzzy genomes. Artif Intell Med 18(1):1–28. https://doi.org/10.1016/s0933-3657(99)00032-9
Kong S-G, Kosko B (1992) Adaptive fuzzy systems for backing up a truck-and-trailer. IEEE Trans Neural Networks 3(2):211–223. https://doi.org/10.1109/72.125862
Qi X et al (2011) A novel model for DNA sequence similarity analysis based on graph theory. Evolut Bioinformatics 7:EBO-S7364. https://doi.org/10.4137/EBO.S7364
Das S et al (2020) A new graph-theoretic approach to determine the similarity of genome sequences based on nucleotide triplets. Genomics 112(6):4701–4714. https://doi.org/10.1016/j.ygeno.2020.08.023
Das S et al (2018) Optimal choice of k-mer in composition vector method for genome sequence comparison. Genomics 110(5):263–273. https://doi.org/10.1016/j.ygeno.2017.11.003
Afreixo V et al (2009) Genome analysis with inter-nucleotide distances. Bioinformatics 25(23):3064–3070. https://doi.org/10.1093/bioinformatics/btp546
Tavares A et al. Detection of exceptional genomic words: a comparison between species. No. 63. EasyChair, 2018.
Tavares H et al (2017) DNA word analysis based on the distribution of the distances between symmetric words. Sci Rep 7(1):728
Goldberger AL, Peng CK (2005) Genomic classification using an information-based similarity index: application to the SARS coronavirus. J Comput Biol 12(8):1103–1116. https://doi.org/10.1089/cmb.2005.12.1103
Pham TD, Zuegg J (2004) A probabilistic measure for alignment-free sequence comparison. Bioinformatics 20(18):3455–3461. https://doi.org/10.1093/bioinformatics/bth426
Kullback S (1968) Information theory and statistics. Dover Publi Inc, New York
Jeffreys H (1946) An invariant form for the prior probability in estimation problems. Proce Royal Soc London Series A Math Phys Sci 186(1007):453–461
Kullback S, Leibler RA (1951) On information and sufficiency. Ann Math Stat 22(1):79–86
Lin J (1991) Divergence measures based on the Shannon entropy. IEEE Trans Inf Theory 37(1):145–151. https://doi.org/10.1109/18.61115
Lu J, Henchion M, MacNamee B. Extending jensen shannon divergence to compare multiple corpora. InMcAuley, J., McKeever, S.(eds.). Proceedings of the 25th Irish Conference on Artificial Intelligence and Cognitive Science 2017. CEUR-WS. org..
Lu G (2013) A class of new metrics for n-dimensional unit hypercube. J Appl Math. https://doi.org/10.1155/2013/942687
Das S et al (2013) Some anomalies in the analysis of whole genome sequence on the basis of Fuzzy set theory. Int J Artif Intell Neural Netw 3(2):38–41
Ghosh S et al (2023) A method of genome sequence comparison based on a new form of fuzzy polynucleotide space Frontiers of ICT in Healthcare. Proceedings of EAIT 2022. Springer Nature Singapore, Singapore, pp 125–135
Saitou N, Nei M (1987) The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425. https://doi.org/10.1093/oxfordjournals.molbev.a040454
Yu C, Deng M, Yau SS (2011) DNA sequence comparison by a novel probabilistic method. Information Sci 181(8):1484–1492. https://doi.org/10.1016/j.ins.2010.12.010
Robinson DF, Foulds LR (1981) Comparison of phylogenetic trees. Math Biosci 53(1–2):131–147
Felsenstein, J. (2005). PHYLIP (phylogeny inference package) Distributed by the author. Dept. Genome Sci., Univ. Wash., Seattle Version, 3.
Author information
Authors and Affiliations
Contributions
SG: Design and development of the work and finalization of draft. JP: Data collection, analysis and interpretation. BM: Initial drafting the article. CC: Critical revision of the article after final draft. DKB: Concepttion of the work and critical revision of the article after final draft.
Corresponding author
Ethics declarations
Competing Interests
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Ghosh, S., Pal, J., Maji, B. et al. Choice of Metric Divergence in Genome Sequence Comparison. Protein J 43, 259–273 (2024). https://doi.org/10.1007/s10930-024-10189-x
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10930-024-10189-x