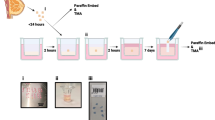
Abstract
Tumor spheroid models have proven useful in the study of cancer cell responses to chemotherapeutic compounds by more closely mimicking the 3-dimensional nature of tumors in situ. Their advantages are often offset, however, by protocols that are long, complicated, and expensive. Efforts continue for the development of high-throughput assays that combine the advantages of 3D models with the convenience and simplicity of traditional 2D monolayer methods. Herein, we describe the development of a breast cancer spheroid image cytometry assay using T47D cells in Aggrewell™400 spheroid plates. Using the Celigo® automated imaging system, we developed a method to image and individually track thousands of spheroids within the Aggrewell™400 microwell plate over time. We demonstrate the use of calcein AM and propidium iodide staining to study the effects of known anti-cancer drugs Doxorubicin, Everolimus, Gemcitabine, Metformin, Paclitaxel and Tamoxifen. We use the image cytometry results to quantify the fluorescence of calcein AM and PI as well as spheroid size in a dose dependent manner for each of the drugs. We observe a dose-dependent reduction in spheroid size and find that it correlates well with the viability obtained from the CellTiter96® endpoint assay. The image cytometry method we demonstrate is a convenient and high-throughput drug-response assay for breast cancer spheroids under 400 μm in diameter, and may lay a foundation for investigating other three-dimensional spheroids, organoids, and tissue samples.





Similar content being viewed by others
Data Availability
The datasets generated during and/or analyzed during the current study are available from the first author on reasonable request.
Code Availability
Not applicable.
References
Selby M et al (2017) 3D Models of the NCI60 Cell Lines for Screening Oncology Compounds. SLAS Discov 22:473–483
Close DA et al (2018) Implementation of the NCI-60 Human Tumor Cell Line Panel to Screen 2260 Cancer Drug Combinations to Generate >3 Million Data Points Used to Populate a Large Matrix of Anti-Neoplastic Agent Combinations (ALMANAC) Database. SLAS DISCOVERY: Advancing the Science of Drug Discovery 24:242–263
Fang Y, Eglen RM (2017) Three-dimensional cell cultures in drug discovery and development. Slas discovery: Advancing Life Sciences R&D 22:456–472
Sant S, Johnston PA (2017) The production of 3D tumor spheroids for cancer drug discovery. Drug Discov Today Technol 23:27–36
Riedl A et al (2017) Comparison of cancer cells in 2D vs 3D culture reveals differences in AKT–mTOR–S6K signaling and drug responses. J Cell Sci 130:203–218
Mirab F, Kang YJ, Majd S (2019) Preparation and characterization of size-controlled glioma spheroids using agarose hydrogel microwells. PLoS ONE 14:e0211078–e0211078
Mortensen ACL, Morin E, Brown CJ, Lane DP, Nestor M (2020) Enhancing the therapeutic effects of in vitro targeted radionuclide therapy of 3D multicellular tumor spheroids using the novel stapled MDM2/X-p53 antagonist PM2. EJNMMI Res 10:38–38
Ballangrud ÅM et al (2001) Response of LNCaP Spheroids after Treatment with an α-Particle Emitter (<sup>213</sup>Bi)-labeled Anti-Prostate-specific Membrane Antigen Antibody (J591). Can Res 61:2008–2014
Sirenko O et al (2015) High-Content Assays for Characterizing the Viability and Morphology of 3D Cancer Spheroid Cultures. Assay Drug Dev Technol 13:402–414
LaBonia GJ, Lockwood SY, Heller AA, Spence DM, Hummon AB (2016) Drug penetration and metabolism in 3D cell cultures treated in a 3D printed fluidic device: assessment of irinotecan via MALDI imaging mass spectrometry. Proteomics 16:1814–1821
Liu X, Weaver EM, Hummon AB (2013) Evaluation of therapeutics in three-dimensional cell culture systems by MALDI imaging mass spectrometry. Anal Chem 85:6295–6302
Hagemann J et al (2017) Spheroid-based 3D Cell Cultures Enable Personalized Therapy Testing and Drug Discovery in Head and Neck Cancer. Anticancer Res 37:2201–2210
Palubeckaitė I et al (2020) Mass spectrometry imaging of endogenous metabolites in response to doxorubicin in a novel 3D osteosarcoma cell culture model. J Mass Spectrom. 55:e4461
Vinci M, Box C, Eccles SA (2015) Three-Dimensional (3D) Tumor Spheroid Invasion Assay. JoVE. e52686 https://doi.org/10.3791/52686
Vinci M, Box C, Zimmermann M, Eccles S (2013) Tumor Spheroid-Based Migration Assays for Evaluation of Therapeutic Agents. Methods Mol Biol 986:253–266
Scherliess R (2011) The MTT assay as tool to evaluate and compare excipient toxicity in vitro on respiratory epithelial cells. Int J Pharm 411:98–105
Larsson P et al (2020) Optimization of cell viability assays to improve replicability and reproducibility of cancer drug sensitivity screens. Sci Rep 10:5798
Eilenberger C et al (2018) Optimized alamarBlue assay protocol for drug dose-response determination of 3D tumor spheroids. MethodsX 5:781–787
Lang SH, Sharrard RM, Stark M, Villette JM, Maitland NJ (2001) Prostate epithelial cell lines form spheroids with evidence of glandular differentiation in three-dimensional Matrigel cultures. Br J Cancer 85:590–599
Dadgar N et al (2020) A microfluidic platform for cultivating ovarian cancer spheroids and testing their responses to chemotherapies. Microsyst Nanoeng 6:93
Ware MJ et al (2016) Generation of Homogenous Three-Dimensional Pancreatic Cancer Cell Spheroids Using an Improved Hanging Drop Technique. Tissue Eng Part C Methods 22:312–321
Ingram M et al (1997) Three-dimensional growth patterns of various human tumor cell lines in simulated microgravity of a NASA bioreactor. In Vitro Cell Dev Biol Anim 33:459–466
Howes AL, Richardson RD, Finlay D, Vuori K.(2014) 3-Dimensional culture systems for anti-cancer compound profiling and high-throughput screening reveal increases in EGFR inhibitor-mediated cytotoxicity compared to monolayer culture systems. PloS One. 9(9):e108283
Vinci M et al (2012) Advances in establishment and analysis of three-dimensional tumor spheroid-based functional assays for target validation and drug evaluation. BMC Biol 10:29
Lee JM et al (2018) Generation of uniform-sized multicellular tumor spheroids using hydrogel microwells for advanced drug screening. Sci Rep 8:1–10
Jeon O, Marks R, Wolfson D, Alsberg E (2016) Dual-crosslinked hydrogel microwell system for formation and culture of multicellular human adipose tissue-derived stem cell spheroids. Journal of Materials Chemistry B 4:3526–3533
Fukuda J et al (2006) Micromolding of photocrosslinkable chitosan hydrogel for spheroid microarray and co-cultures. Biomaterials 27:5259–5267
Okuyama T et al (2010) Preparation of arrays of cell spheroids and spheroid-monolayer cocultures within a microfluidic device. J Biosci Bioeng 110:572–576
Seo J et al (2018) High-throughput approaches for screening and analysis of cell behaviors. Biomaterials 153:85–101
Sun Q et al (2018) Microfluidic Formation of Coculture Tumor Spheroids with Stromal Cells As a Novel 3D Tumor Model for Drug Testing. ACS Biomater Sci Eng 4:4425–4433
Madoux F et al (2017) A 1536-Well 3D Viability Assay to Assess the Cytotoxic Effect of Drugs on Spheroids. SLAS DISCOVERY: Advancing the Science of Drug Discovery 22:516–524
Mathews LA et al (2012) A 1536-well quantitative high-throughput screen to identify compounds targeting cancer stem cells. J Biomol Screen 17:1231–1242
Tung Y-C et al (2011) High-throughput 3D spheroid culture and drug testing using a 384 hanging drop array. Analyst 136:473–478
Raghavan S et al (2015) Formation of stable small cell number three-dimensional ovarian cancer spheroids using hanging drop arrays for preclinical drug sensitivity assays. Gynecol Oncol 138:181–189
Razian G, Yu Y, Ungrin M (2013) Production of large numbers of size-controlled tumor spheroids using microwell plates. JoVE J Visual Exp. 81:e50665. https://doi.org/10.3791/50665
Chan LL, Wilkinson AR, Paradis BD, Lai N (2012) Rapid Image-based Cytometry for Comparison of Fluorescent Viability Staining Methods. J Fluoresc 22:1301–1311
Chan LL-Y, Kuksin D, Laverty DJ, Saldi S, Qiu J (2015) Morphological observation and analysis using automated image cytometry for the comparison of trypan blue and fluorescence-based viability detection method. Cytotechnology 67:461–473
Cribbes S, Kessel S, McMenemy S, Qiu J, Chan LL-Y (2017) A Novel Multiparametric Drug-Scoring Method for High-Throughput Screening of 3D Multicellular Tumor Spheroids Using the Celigo Image Cytometer. SLAS DISCOVERY: Advancing the Science of Drug Discovery 22:547–557
Pereira P et al (2017) Cancer cell spheroids are a better screen for the photodynamic efficiency of glycosylated photosensitizers. PLoS ONE. 12
Gaskell H et al (2016) Characterization of a Functional C3A Liver Spheroid Model. Toxicol. Res. 5
Gong X et al (2015) Generation of Multicellular Tumor Spheroids with Microwell-Based Agarose Scaffolds for Drug Testing. Plos One 10:e0130348
Lewis N et al (2017) Magnetically levitated mesenchymal stem cell spheroids cultured with a collagen gel maintain phenotype and quiescence. Journal of Tissue Engineering 8:204173141770442
Singh M, Close DA, Mukundan S, Johnston PA, Sant S (2015) Production of Uniform 3D Microtumors in Hydrogel Microwell Arrays for Measurement of Viability, Morphology, and Signaling Pathway Activation. Assay Drug Dev Technol 13:570–583
Singh M, Mukundan S, Jaramillo M, Oesterreich S, Sant S (2016) Three-Dimensional Breast Cancer Models Mimic Hallmarks of Size-Induced Tumor Progression. Can Res 76:3732–3743
Thakuri PS, Gupta M, Plaster M, Tavana H (2019) Quantitative Size-Based Analysis of Tumor Spheroids and Responses to Therapeutics. Assay Drug Dev Technol 17:140–149
Yang W et al (2012) Genomics of Drug Sensitivity in Cancer (GDSC): a resource for therapeutic biomarker discovery in cancer cells. Nucleic Acids Res 41:D955–D961
Ariaans G, Jalving M, Vries EG, Jong S (2017) Anti-tumor effects of everolimus and metformin are complementary and glucose-dependent in breast cancer cells. BMC Cancer 17:232
Serrano MJ et al (2002) Evaluation of a Gemcitabine-Doxorubicin-Paclitaxel Combination Schedule through Flow Cytometry Assessment of Apoptosis Extent Induced in Human Breast Cancer Cell Lines. Jpn J Cancer Res 93:559–566
Bashmail HA et al (2018) Thymoquinone synergizes gemcitabine anti-breast cancer activity via modulating its apoptotic and autophagic activities. Sci Rep 8:11674
Durbin KR, Nottoli MS, Jenkins GJ (2020) Effects of microtubule-inhibiting small molecule and antibody-drug conjugate treatment on differentially-sized A431 squamous carcinoma spheroids. Sci Rep 10:907
Funding
This research was conducted with support under Grant No. R01EB012521 awarded by the National Institutes of Health.
Author information
Authors and Affiliations
Contributions
SM and JB: Conceptualization; data curation; formal analysis; investigation; methodology; visualization; writing original draft; writing-review and editing. MT: data curation; formal analysis. CH and ACL: Methodology; writing-review and editing. BP and LLC: Project administration; resources; supervision; conceptualization; visualization; writing original draft; writing-review and editing.
Corresponding author
Ethics declarations
Ethics Approval
Not applicable.
Consent to Participate
Not applicable.
Consent for Publication
Not applicable.
Conflict of Interest
The authors JB, CH, ACL, and LLC declare competing financial interests. The tumor spheroid detection method development in this manuscript is for reporting on a biological application using an instrument of Nexcelom Bioscience, LLC. The work demonstrated a high-throughput method for the characterization of tumor spheroids in Aggrewell plates using the Celigo® Image Cytometer.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Mukundan, S., Bell, J., Teryek, M. et al. Automated Assessment of Cancer Drug Efficacy On Breast Tumor Spheroids in Aggrewell™400 Plates Using Image Cytometry. J Fluoresc 32, 521–531 (2022). https://doi.org/10.1007/s10895-021-02881-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10895-021-02881-3