Abstract
Large coupling networks in uniformly 13C,15N-labeled biomolecules induce broad multiplets that even in flexible proteins are frequently not recognized as such. The reason is that given multiplets typically consist of a large number of individual resonances that result in a single broad line, in which individual components are no longer resolved. We here introduce a real-time pure shift acquisition scheme for the detection of amide protons which is based on 13C-BIRDr,X. As a result the full homo- and heteronuclear coupling network can be suppressed at low power leading to real singlets at substantially improved resolution and uncompromised sensitivity. The method is tested on a small globular and an intrinsically disordered protein (IDP) where the average spectral resolution is increased by a factor of ~ 2 and higher. Equally important, the approach works without saturation of water magnetization for solvent suppression and exchanging amide protons are not affected by saturation transfer.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Modern NMR spectroscopy is an indispensable tool for obtaining valuable information at atomic resolution. However, this information may be obliterated if the studied biomolecules contain repeating units, or they belong to the group of intrinsically disordered proteins (IDPs)—where chemical shifts are distributed close to random coil values and very little dispersion is experienced (Dyson and Wright 2001). Large efforts have been made to overcome signal overlap, either by performing experiments at higher fields, by applying detection of heteronuclei (Serber et al. 2000; Bermel et al. 2006, 2008, 2012; Felli and Pierattelli 2014; Chhabra et al. 2018) or by introducing experiments of high dimensionality (Bermel et al. 2009; Narayanan et al. 2010; Bertini et al. 2011). Another possibility is offered by so-called pure shift methods that allow the collapse of the broad, sometimes unresolved multiplets to a single line—which has the advantage to increase both sensitivity and resolution for fast tumbling proteins. However, from the many available pure shift methods (Zangger 2015; Castañar and Parella 2015; Hammarström and Otting 1994; Kupče and Wagner 1995; Castañar et al. 2013; Ying et al. 2014; Kiraly et al. 2015; Im et al. 2019; Haller et al. 2019; Bodor et al. 2020; Meyer and Zangger 2014, 2013; Zangger and Sterk 1997; Nilsson and Morris 2007; Sakhaii et al. 2009; Aguilar et al. 2011; Lupulescu et al. 2012; Paudel et al. 2013; Reinsperger and Luy 2014; Foroozandeh et al. 2014; Sinnaeve et al. 2016) only the ones with essentially uncompromised sensitivity may be applicable to biomolecules (Hammarström and Otting 1994; Kupče and Wagner 1995; Castañar et al. 2013; Ying et al. 2014; Kiraly et al. 2015; Im et al. 2019; Haller et al. 2019; Bodor et al. 2020; Meyer and Zangger 2014).
Since detection of protons provides highest sensitivity, it is well-established in most standard experiments of the field (Clore and Gronenborn 1991). In particular amide protons are widely used for detection, and it is worth to address the special challenges for their pure shift acquisition in uniformly 13C,15N-labeled samples. A drawback of this approach is that amide protons being in exchange with water imply the presence of an enormous solvent signal that has to be suppressed during the NMR experiment. Solvent suppression has turned out to be especially challenging for pure shift methods where pulses are applied during the acquisition and water magnetization so far had to be saturated using a weak radiofrequency field (rf) or pulsed field gradients (Castañar et al. 2013; Ying et al. 2014; Kiraly et al. 2015). As a consequence, amide protons in exchange with the solvent suffer from signal attenuation (Grzesiek and Bax 1993; Mori et al. 1995) which is especially striking for IDPs studied close to neutral pH (Bai et al. 1993).
Furthermore, in uniformly 13C,15N-labeled biomolecules a large number of homo- and heteronuclear couplings are encountered that, even if unresolved, cause signal broadening and highest resolution can only be obtained if all present couplings are suppressed. Up to now, the suppression of numerous heteronuclear long-range couplings is achieved by using power-intensive composite pulse decoupling (CPD), sometimes even on both heteronuclei, at high strain for probe and hardware. This poses a significant limit to the acquisition time and hence to the achievable resolution (Ying et al. 2014).
Here, we present an acquisition scheme for the pure shift detection of amide protons in uniformly 13C,15N-labeled proteins, applicable both to small globular proteins and IDPs, that overcomes the mentioned issues. The method is based on the real-time decoupling approach as originally proposed by Lupulescu et al. (Lupulescu et al. 2012). The central element of the decoupling sequence is a 13C-BIRDr,X filter (Garbow et al. 1982; Uhrín et al. 1993) that enables suppression of all small homo- and heteronuclear couplings and avoids the need of 13C-CPD (Haller et al. 2019; Bodor et al. 2020). As a result, uncompromised acquisition times are accessible and high resolution is achieved with only 15N-decoupling to be considered if applicable. Moreover, water polarization can be stored along the z-axis during acquisition—avoiding the need of saturation of the water resonance and allowing the water spin reservoir to be used for fast pulsing experiments (Schanda 2009).
We here use the proposed pure shift acquisition in simple FHSQC (Mori et al. 1995) and BEST-TROSY (Solyom et al. 2013) measurements, but it should be applicable in any 1H,15N-HSQC or TROSY-type experiment. Results are shown for the small globular protein ubiquitin, and for the intrinsically disordered protein domain p53TAD1−60, where a significant enhancement in resolution can be observed at high sensitivity.
The pulse sequence
Modern real-time homonuclear decoupling requires a selective inversion element that allows to distinguish the detected active spins from their passive coupling partners and can hence act as J-refocusing block—a pure shift spectrum can then be acquired in a single shot (Lupulescu et al. 2012). For this purpose, the acquisition is repeatedly interrupted and the FID is recorded in chunks. Each time the acquisition is paused, couplings to passive spins are refocused by the selective inversion element while acquired active spins are in total not affected. If the chunk length is chosen short compared to the inverse multiplet width, passive couplings cannot evolve significantly during acquired chunks and decoupling is thus achieved to a good approximation. For the detection of amide protons, the discrimination between active and passive spins can be obtained e.g., with band-selective pulses (Castañar et al. 2013; Ying et al. 2014) or using a 15N-BIRD filter (Kiraly et al. 2015).
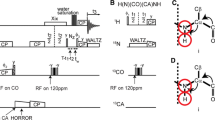
For uniformly 13C,15 N-labeled proteins, we propose the approach shown in Fig. 1. In the given pure shift acquisition sequence (Fig. 1b), the selective inversion element consists of a 13C-BIRDr,X filter in combination with a hard 180° pulse on the proton channel. Following the nomenclature of Uhrín et al., the BIRD-filter selectively inverts remote protons (r) and carbons (X), while protons directly bound to 13C (d) are not inverted (Uhrín et al. 1993). For clarity, the pulse sequence is again shown in Fig. 1c using a pseudo pulse sequence where the BIRD-filter is reduced to the effective pulses of the element for remote (1Hr) and directly 13C-bound protons (1Hd). It is obvious that acquired amide protons (as part of 1Hr) experience two inversions and, if relaxation and pulse imperfections are neglected, they are thus not affected by the selective inversion element. On the other hand, all carbons as well as protons directly attached to 13C (1Hd) experience only one effective inversion. Therefore, all coupling partners of the amide protons are inverted by the J-refocusing element (except 15N) leading to the real-time suppression of both homonuclear 1Hr,1Hd-couplings as well as heteronuclear long-range 1Hr,13C-couplings. Carbon decoupling is hence achieved at minimal additional cost of power and long acquisition times are not restricted by the 13C-decoupler. Note that nearly all 2J- and 3J-couplings of the amide proton are heteronuclear (Fig. 1a) and in total, the numerous couplings can cause substantial broadening. In 1H,15N-HSQC-type experiments only the large 1JNH coupling additionally needs to be suppressed by 15N-CPD during detection. In TROSY-type experiments, on the other hand, the 1JNH coupling is supposed to evolve and, practically, there is no rf-limit imposed on the acquisition time.
a Relevant J-couplings for amide protons (Salvador et al. 2011). b Pulse sequence for real-time decoupled acquisition of amide protons using a 13C-BIRDr,X filter (blue dashed box) (Garbow et al. 1982; Uhrín et al. 1993). CPD on 15N (in grey) is applied for 1H,15N HSQC-type experiments, but omitted for TROSY-type experiments. Filled narrow and open wide bars correspond to 90° and 180° pulses, respectively, with phase x if not denoted otherwise. A broadband BIP or BIBOP inversion pulse (I) (Kobzar et al. 2004, 2008; Ehni and Luy 2013; Smith et al. 2001; Baum et al. 1985) covering ~ 200 ppm is applied with a pulse length of 500 μs. ∆ and δ are set to 1/(2 × 1JCαH) and 0.5 ms, respectively. Gradients are used to remove artifacts (G1 ≈ 3.0%) and to avoid radiation damping (G2 ≈ 0.1%). In order to achieve acquisition times of > 200 ms, the number of chunks is typically set to n = 8 with half chunk lengths of tc = 14–18 ms and breaks of tb ≈ 9.2 ms. c The effective rotations of the BIRDr,X element are illustrated for directly 13C-bound (Hd, red) and remote (Hr, blue) protons.
Furthermore, it is crucial to note that also water belongs to the group of remote protons (1Hr) and, to good extent, it is likewise not affected by the selective inversion element which has remarkable consequences in terms of solvent suppression.
Water suppression
The large excess of H2O in biomolecular samples prevents the detection of much smaller protein peaks and the water signal intensity has to be reduced drastically. Saturation of the water bulk magnetization from a weak rf-field causes signal attenuation of exchanging amide protons and should hence be avoided (Mori et al. 1995; Hoult 1976). Note, using pulsed field gradients to dephase the water signal can also lead to saturation of amide protons—typical T1 relaxation times of water are in the range of several seconds, which is much longer than conventional recovery delays. In order to avoid saturation transfer to exchanging protons (and consecutively also to surrounding spins via NOE and spin diffusion) it is thus advisable to apply so-called flip-back techniques that store the water bulk magnetization along the non-observable + z axis during acquisition (Jahnke and Kessler 1994; Lippens et al. 1995). The bulk water can thereby be used as a large spin reservoir for exchanging protons in fast pulsing experiments that in favorable cases allow a vast reduction of the recovery delay (tr) (Mori et al. 1995; Solyom et al. 2013).
With respect to real-time homonuclear decoupling, one should be aware that already a small perturbation to the strong water resonance could cause enormous artifacts. For this reason, common real-time pure shift sequences make use of pulsed field gradients, which are capable to suppress transverse magnetization that originates from pulse imperfections during the chunked acquisition.
If, however, the water bulk magnetization has to be flipped by the selective inversion element from the + z to the − z axis, radiation damping will cause artifact buildup during detection where pulsed field gradients are not applicable and the strong water signal will reappear. The emerging artifacts in such a case are enormous and are shown for a pure shift FHSQC using 15N-BIRD in Fig. 2a and aʹ.
Water artifacts caused by radiation damping in a pure shift FHSQC using a 15N-BIRD (left) and 13C-BIRDr,X filter (right). Only the first FID was acquired with 16 dummy scans using a cryogenically cooled TCI probehead (a, b). Blue areas indicate where water bulk magnetization is flipped to − z axis with strong radiation damping. Respective spectra with resulting artifacts are shown below (aʹ, bʹ)
In every second chunk, water is flipped to the − z axis where radiation damping non-linearly rotates the water magnetization and the entire 1D spectrum is dominated by the water chunking artifact. Note that also the use of band-selective homonuclear decoupling would lead to comparable results.
On the other hand, with our proposed sequence using a 13C-BIRDr,X filter, the water bulk magnetization can be kept along the + z axis and artifacts due to radiation damping can be reduced by two orders of magnitude as shown in Fig. 2b and bʹ. By this means, the quality of water suppression is on an equal level compared to other pure shift methods (see Fig. S1) while the water bulk magnetization can be retained for exchanging amide protons. Bear in mind, that radiation damping strongly depends on the employed probe and the quality of water suppression can vary significantly for different hardware and settings. For best water suppression we recommend to follow the procedure described in the supporting information.
Materials and methods
NMR spectra were recorded on a 600 MHz Bruker Avance III spectrometer equipped with a 5 mm H-C/N-D TCI cryogenically cooled probehead and a 16.4 T Bruker Avance III spectrometer operating at 700.05 MHz equipped with a 5 mm Prodigy H&F-C/N-D TCI probehead. The composition of the U-13C/15N labeled protein samples was: 0.5 mM human ubiquitin by Silantes in sealed 5 mm NMR tube, 7% D2O, pH 4.7, 30 mM sodium acetate, 50 mM NaCl; 1 mM p53TAD1−60, 20 mM MES (2-(N-morpholino)ethanesulfonic acid), 20 mM NaCl, 10 mM TCEP (tris (2-carboxyethyl)phosphine), 10% D2O, pH 6.0, prepared as described earlier (Dudas et al. 2020).
Application to proteins
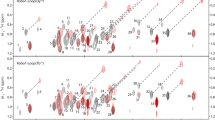
Due to the sheer number of amide proton couplings in uniformly 13C,15N-labeled proteins, the splittings contribute significantly to the linewidths of small globular or intrinsically disordered proteins, where natural linewidths are expected to be relatively narrow. In many such cases, unresolved multiplets likely dominate the actual signal width and pure shift methods may lead to an increase in spectral resolution and sensitivity. The first example is given in Fig. 3 for the 76 residue long folded ubiquitin, where homo- and long-range heteronuclear couplings are removed using the 13C-BIRD-based pure shift acquisition in an FHSQC and BEST-TROSY. A closer analysis of the highlighted slices through selected cross peaks shows that signals obtained by standard detection exhibit a very broad top of the signal that indicates the presence of multiplets. If these underlying multiplets are collapsed, sharp Lorentzian-type singlets are obtained. As seen in Fig. 3, the standard acquisition scheme results in signal widths of 10–23 Hz for both FHSQC and TROSY spectra, with slightly narrower lines for the TROSY as expected. Introduction of the 13C-BIRDr,X-based pure shift acquisition (aʹ, bʹ) results in considerable narrowing to approximately half the signal width. The main width reduction is due to the suppression of 1HN,1H-couplings and it should be noted that also long-range 1HN,13C-couplings contribute significantly to line broadening. Suppressing 1HN,13C-couplings in case of ubiquitin we find an average reduction in signal width of 3–4 Hz (Fig. S2). Since decoupling of long-range 1HN,13C-couplings consumes negligible additional power, the achievable resolution is not hardware-limited and for the discussed experiments acquisition times up to 321 ms were used. Moreover, only with full decoupling the favorable relaxation properties of TROSY are revealed. Despite the fact that cross-correlated relaxation is more pronounced for larger proteins, already for ubiquitin we find a considerable increase in resolution using TROSY and overlapping signals as e.g. Q31 and R72 can be resolved unambiguously as highlighted in Fig. 3bʹ.
Spectra of ubiquitin acquired at 600 MHz using FHSQC (left) and BEST-TROSY (right) with standard acquisition (a, b) and 13C-BIRDr,X-based homonuclear decoupling (aʹ, bʹ). While in the FHSQC 2048 complex points were acquired in 214 ms with tc = 17.8 ms and n = 6, for TROSY 3096 complex points were recorded in 321 ms with tc = 20 ms and n = 8 using a magnetization recovery delay of tr = 1.0 s. The assignments are based on literature (Weber et al. 1987; Wang et al. 1995) and overlapping signals (Q31 and R72) can be resolved in (aʹ, bʹ). Correlation plots are shown for the FHSQC and BEST-TROSY in (aʹʹ, bʹʹ), respectively, where signal widths with standard acquisition and homonuclear decoupling are compared. Dashed lines mark average values and a quadratic phase-shifted sine (a, b, aʹ, bʹ) and no apodization (aʹʹ, bʹʹ) was used for processing
To provide a wider estimate on the increase in resolution, signal widths at half height for spectra with and without the proposed pure shift sequence were extracted using a semi-automated python script. Correlated values are illustrated in Fig. 3aʹʹ and bʹʹ. In these correlation plots, based on spectra without apodization, we find a reduction in signal width from average values (dashed lines) of 16.3 Hz to 8.8 Hz for the FHSQC and from 16.0 to 7.3 Hz for BEST-TROSY when decoupling is applied. Note that a certain line broadening is induced by quadratic phase-shifted sine apodization and average line widths are increased to 10.7 Hz and 8.3 Hz for the pure shift FHSQC and BEST-TROSY, respectively. Improved linewidths can be obtained using processing parameters as described elsewhere (Kiraly et al. 2018).
Based on the fact that transverse relaxation is typically much slower for intrinsically disordered proteins, in this case even sharper lines are expected using the 13C-BIRDr,X pure shift method. In Fig. 4 we show the result obtained from BEST-TROSY measurements for the disordered p53TAD1−60 (Dudas et al. 2020). To obtain high resolution, a relatively long acquisition time of 243 ms was chosen.
Spectra of p53TAD1-60 were acquired at 700 MHz using BEST-TROSY with standard acquisition (a) and homonuclear decoupling (b). 2048 complex points were acquired in 243 ms with tc = 15.2 ms and n = 8 at a temperature of 298 K and a recovery delay of tr = 200 ms. Signal widths are compared using correlation plots (c) where dashed lines mark average values and no apodization was used for processing in (a–c)
As can be seen in the highlighted 1D slices, a substantial enhancement in resolution is obtained. The corresponding correlation plot (Fig. 4c) reveals that the average signal width of 12.7 Hz for standard acquisition can be reduced to as low as 5.0 Hz using the 13C-BIRD-based pure shift sequence. Such a significant enhancement in resolution by a factor of ~ 2.5 is of major relevance for IDPs where overcrowded spectra are not only caused by low signal dispersion but frequently also by additional resonances originating from minor conformations.
From the collapse of multiplets one would expect that signal intensities are increased accordingly—the influence on sensitivity using pure shift, is, however, more complex. Both, the FHSQC and BEST-TROSY experiments make use of spin reservoirs that need to persist during the entire sequence and in turn allow high repetition rates. If such sensitivity-trimmed experiments are applied in combination with the 13C-BIRD-based pure shift acquisition, certain aspects with respect to sensitivity have to be reconsidered: (i) pure shift methods are committed to highest resolution—long acquisition times are hence mandatory, which dampens experimental repetition rates and should also be considered when using CPD (in HSQC) to avoid hardware damage; (ii) to a certain extent magnetization recovery of amide protons from longitudinal relaxation is hindered by repetitive pulsing during pure shift acquisition; (iii) an accelerated build-up of magnetization via NOE and spin diffusion (BEST approach) is not expected during the acquisition since the spin reservoir of surrounding protons is repeatedly flipped—an even number of chunks might partially retain the effect during the recovery delay, but particularly the recovery of reservoir polarization during acquisition does not take place; (iv) a faster magnetization build-up using the water spin reservoir is still possible, while, on the other hand, solvent exchange might induce line broadening which lowers the achievable resolution; (v) incomplete water suppression might enhance artificial noise originating from the chunked acquisition; (vi) the collapse of broad unresolved multiplets increases signal intensities to an extent that before-mentioned effects may well be compensated. Based on all these aspects, it is rather difficult to quantify the influence on sensitivity obtained for a general case when using the 13C-BIRD-based pure shift acquisition.
However, one can estimate the increase in sensitivity obtained by the pure shift FHSQC from Fig. 5 where standard and pure shift ubiquitin spectra are analyzed using four different recovery delays \({t}_{r}\) (ranging from 0.8 to 1.5 s not including acquisition times). In all spectra, maximum intensities of amide proton signals are extracted and average values are shown for standard (blue) and pure shift acquisition (red). The averaged data is fitted to mono-exponential recovery functions with \(I ({t}_{r})={I}_{E}\cdot (1-{I}_{0}\cdot {e}^{-{R}_{1}{t}_{r}})\) where \({I}_{E}\) corresponds to the equilibrium magnetization, \({I}_{0}\) is the magnetization for \({t}_{r}=0\) and \({R}_{1}\) is the fitted average longitudinal relaxation rate. Note, repetitive pulsing interferes with magnetization buildup during real-time chunked acquisition and a lower value of \({I}_{0}\) is obtained for pure shift detection. From the fitting, it is hence possible to conclude that for very short recovery delays on average higher maximum intensities are found using standard acquisition (blue). Yet, already for recovery delays of tr > 250 ms the gain in sensitivity due to collapsing multiplets outweighs the sensitivity of conventional acquisition and maximum intensities are on average increased by up to 32% using the 13C-BIRD-based pure shift detection. Note, total experimental times are increased using the decoupling scheme as shown in Table S1.
Discussion
In the past decade various methods have been proposed for the real-time chunked acquisition of protein amide protons with which a significant enhancement in resolution can be achieved (Castañar et al. 2013; Ying et al. 2014; Kiraly et al. 2015). While in uniformly 15N-labeled proteins it is sufficient to suppress homonuclear 1HN,1H-couplings using band-selective or 15N-BIRD-based pure shift detection, numerous heteronuclear 1HN,13C-couplings cause significant line broadening if also 13C-spin labelling is applied. The straightforward use of 13C-CPD will lead to a dramatic increase in applied rf-energy during acquisition and appears exaggerated in isotropic liquids, where heteronuclear long-range couplings are almost two orders of magnitude smaller than typical 1JCH couplings. It is hence adequate to include carbon decoupling in the real-time pure shift detection and high resolution can be obtained practically without hardware-limitation. Including the suppression of heteronuclear long-range couplings in the 13C-BIRD-based detection, we find that half chunk lengths (tc) of 15–20 ms are suitable to avoid considerable coupling evolution while keeping the interruptive nature of chunked acquisitions at a minimum. Possible small chunking sidebands from residual J-evolution may be suppressed by frequency or phase modulation, which is appropriate e.g. for IDPs where minor conformations induce additional sets of resonances with reduced intensities (Mauhart et al. 2015; Moutzouri et al. 2017).
It is further worth mentioning that transverse relaxation during chunking breaks (tb) introduces an artificial decay and the achievable line width is increased by a factor of \((2{t}_{c}+{t}_{b})/ (2{t}_{c})\) (Ying et al. 2014). For this reason, shorter breaks tb are advantageous and the artificial decay induced by the 13C-BIRD-based acquisition (tb ≈ 9.2 ms) is less pronounced than with the 15N-BIRD filter (tb ≈ 13.2 ms). The shortest interruption is, however, found using optimized band-selective pulses (tb ≈ 3–5 ms) and highest resolution would be obtained if it were not for the long-range heteronuclear couplings. For relaxation-limited samples hence, a subtle approach combining BASHD/HOBS with broadband 13C-inversions simultaneously applied to band-selective pulses might lead to the best compromise, which we, however, did not implement on our available, clearly more multiplet-broadened test samples. It is, however, crucial to note that the band-selective approach will fail for exchanging amide protons, since solvent suppression would require the saturation of the water resonance, thereby also removing corresponding amide signals.
Concluding remarks
A novel selective element based on 13C-BIRDr,X inversion for the real-time pure shift acquisition of amide protons in uniformly 13C,15N-labeled proteins has been introduced. An advantage of the method is, that numerous long-range 1HN,13C-couplings are suppressed without applying rf-intensive 13C-CPD, resulting in only a negligible effect on the duty cycle so that long acquisition times for highest resolution are still achievable. We present the application of this pure shift acquisition scheme incorporated in FHSQC and BEST-TROSY measurements for small folded proteins and IDPs. Experimental data reveal that on average the spectral resolution is increased by a factor of ~ 2 for the folded ubiquitin and ~ 2.5 for the IDP p53TAD1-60, respectively, while the average signal intensity in our example spectra was increased by approximately 30%. As saturation of water is not needed for solvent suppression we showed that the proposed pure shift sequence is perfectly suitable for amide proton detection of proteins. The obtained well-resolved 2D spectra are of great help in automated peak picking, and promotes a more straightforward assignment of overlapping peaks. The incorporation of the 13C-BIRDr,X acquisition scheme into 3D and higher dimensional experiments should be straight-forward. It is especially suitable for 13C,15N-isotope-labeled IDPs where low natural linewidths allow a remarkable advantage from collapsing multiplets while solvent exchange needs to be considered for amide protons.
Data availability
The pulse sequences, raw and processed data will be available upon request.
References
Aguilar JA, Nilsson M, Morris GA (2011) Simple proton spectra from complex spin systems: pure shift NMR spectroscopy using BIRD. Angew Chem Int Ed 50:9716–9717
Bai Y, Milne JS, Mayne L, Englander SW (1993) Primary structure effects on peptide group hydrogen exchange. Proteins 17:75–86
Baum J, Tycko R, Pines A (1985) Broadband and adiabatic inversion of a two-level system by phase-modulated pulses. Phys Rev 32:3435–3447
Bermel W, Bertini I, Felli IC, Kümmerle R, Pierattelli R (2006) Novel 13C direct detection experiments, including extension to the third dimension, to perform the complete assignment of proteins. J Magn Reson 178:56–64
Bermel W, Felli IC, Kümmerle R, Pierattelli R (2008) 13C direct-detection biomolecular NMR. Concepts Magn Reson 32A:183–200
Bermel W, Bertini I, Felli IC, Pierattelli R (2009) Speeding up 13C direct detection biomolecular NMR spectroscopy. J Am Chem Soc 131:15339–15345
Bermel W, Bertini I, Chill J, Felli IC, Haba N, Kumar MVV, Pierattelli R (2012) Exclusively heteronuclear 13C-detected amino-acid-selective NMR experiments for the study of intrinsically disordered proteins (IDPs). ChemBioChem 13:2425–2432
Bertini I, Felli IC, Gonnelli L, Vasantha Kumar MV, Pierattelli R (2011) High-resolution characterization of intrinsic disorder in proteins: expanding the suite of 13C-detected NMR spectroscopy experiments to determine key observables. ChemBioChem 12:2347–2352
Bodor A, Haller JD, Bouguechtouli C, Theillet FX, Nyitray L, Luy B (2020) Power of pure shift HɑCɑ correlations: a way to characterize biomolecules under physiological conditions. Anal Chem 92:12423–12428
Castañar L, Parella T (2015) Broadband 1H homodecoupled NMR experiments: recent developments, methods and applications. Magn Reson Chem 53:399–426
Castañar L, Nolis P, Virgili A, Parella T (2013) Full sensitivity and enhanced resolution in homodecoupled band-selective NMR experiments. Chemistry 19:17283–17286
Chhabra S, Fischer P, Takeuchi K, Dubey A, Ziarek JJ, Boeszoermenyi A, Mathieu D, Bermel W, Davey NE, Wagner G, Arthanari H (2018) 15N detection harnesses the slow relaxation property of nitrogen: delivering enhanced resolution for intrinsically disordered proteins. Proc Natl Acad Sci USA 115:E1710–E1719
Clore GM, Gronenborn AM (1991) Applications of three- and four-dimensional heteronuclear NMR spectroscopy to protein structure determination. Prog Nucl Magn Reson Spectrosc 23:43–92
Dudas EF, Palfy G, Menyhard DK, Sebak F, Ecsedi P, Nyitray L, Bodor A (2020) Tumor-suppressor p53TAD1–60 forms a fuzzy complex with metastasis-associated S100A4: structural insights and dynamics by an NMR/MD Approach. ChemBioChem 21:3087–3095
Ehni S, Luy B (2013) BEBEtr and BUBI: J-compensated concurrent shaped pulses for 1H–13C experiments. J Magn Reson 232:7–17
Felli IC, Pierattelli R (2014) Novel methods based on 13C detection to study intrinsically disordered proteins. J Magn Reson 241:115–125
Foroozandeh M, Adams RW, Meharry NJ, Jeannerat D, Nilsson M, Morris GA (2014) Ultrahigh-resolution NMR spectroscopy. Angew Chem Int Ed 53:6990–6992
Garbow JR, Weitekamp DP, Pines A (1982) Bilinear rotation decoupling of homonuclear scalar interactions. Chem Phys Lett 93:504–509
Grzesiek S, Bax A (1993) The importance of not saturating water in protein NMR. Application to sensitivity enhancement and NOE measurements. J Am Chem Soc 115:12593–12594
Haller JD, Bodor A, Luy B (2019) Real-time pure shift measurements for uniformly isotope-labeled molecules using X-selective BIRD homonuclear decoupling. J Magn Reson 302:64–71
Hammarström A, Otting G (1994) Improved spectral resolution in 1H NMR spectroscopy by homonuclear semiselective shaped pulse decoupling during acquisition. J Am Chem Soc 116:8847–8848
Hoult DI (1976) Solvent peak saturation with single phase and quadrature Fourier transformation. J Magn Reson 21:337–347
Im J, Lee J, Lee JH (2019) Pre-homonuclear decoupling enables high-resolution NMR analysis of intrinsically disordered proteins in solution. J Phys Chem Lett 10:4720–4724
Jahnke W, Kessler H (1994) Enhanced sensitivity of rapidly exchanging amide protons by improved phase cycling and the constructive use of radiation damping. J Biomol NMR 4:735–740
Dyson JH, Wright PE (2001) Nuclear magnetic resonance methods for elucidation of structure and dynamics in disordered states. Methods Enzymol 339:258–270
Kiraly P, Adams RW, Paudel L, Foroozandeh M, Aguilar JA, Timári I, Cliff MJ, Nilsson M, Sandor P, Batta G, Waltho JP, Kövér KE, Morris GA (2015) Real-time pure shift 15N HSQC of proteins: a real improvement in resolution and sensitivity. J Biomol NMR 62:43–52
Kiraly P, Nilsson M, Morris GA (2018) Practical aspects of real-time pure shift HSQC experiments. Magn Reson Chem 56:993–1005
Kobzar K, Skinner TE, Khaneja N, Glaser SJ, Luy B (2004) Exploring the limits of broadband excitation and inversion pulses. J Magn Reson 170:236–243
Kobzar K, Skinner TE, Khaneja N, Glaser SJ, Luy B (2008) Exploring the limits of broadband excitation and inversion: II. Rf-power optimized pulses. J Magn Reson 194:58–66
Kupče Ē, Wagner G (1995) Wideband homonuclear decoupling in protein spectra. J Magn Reson B 109:329–333
Lippens G, Dhalluin C, Wieruszeski JM (1995) Use of a water flip-back pulse in the homonuclear NOESY experiment. J Biomol NMR 5:327–331
Lupulescu A, Olsen GL, Frydman L (2012) Toward single-shot pure-shift solution 1H NMR by trains of BIRD-based homonuclear decoupling. J Magn Reson 218:141–146
Mauhart J, Glanzer S, Sakhaii P, Bermel W, Zangger K (2015) Faster and cleaner real-time pure shift NMR experiments. J Magn Reson 259:207–215
Meyer NH, Zangger K (2013) Simplifying proton NMR spectra by instant homonuclear broadband decoupling. Angew Chem Int Ed 52:7143–7146
Meyer NH, Zangger K (2014) Boosting the resolution of 1H NMR spectra by homonuclear broadband decoupling. ChemPhysChem 15:49–55
Mori S, Abeygunawardana C, Johnson MO, van Zijl PC (1995) Improved sensitivity of HSQC spectra of exchanging protons at short interscan delays using a new fast HSQC (FHSQC) detection scheme that avoids water saturation. J Magn Reson B 108:94–98
Moutzouri P, Chen Y, Foroozandeh M, Kiraly M, Phillips AR, Coombes SR, Nilsson M, Morris GA (2017) Ultraclean pure shift NMR. Chem Commun 53:10188–10191
Narayanan RL, Durr UH, Bibow S, Biernat J, Mandelkow E, Zweckstetter M (2010) Automatic assignment of the intrinsically disordered protein Tau with 441-residues. J Am Chem Soc 132:11906–11907
Nilsson M, Morris GA (2007) Pure shift proton DOSY: diffusion-ordered 1H spectra without multiplet structure. Chem Commun 933-935
Paudel L, Adams RW, Kiraly P, Aguilar JA, Foroozandeh M, Cliff MJ, Nilsson M, Sandor P, Waltho JP, Morris GA (2013) Simultaneously enhancing spectral resolution and sensitivity in heteronuclear correlation NMR spectroscopy. Angew Chem Int Ed 52:11616–11619
Reinsperger T, Luy B (2014) Homonuclear BIRD-decoupled spectra for measuring one-bond couplings with highest resolution: CLIP/CLAP-RESET and constant-time-CLIP/CLAP-RESET. J Magn Reson 239:110–120
Sakhaii P, Haase B, Bermel W (2009) Experimental access to HSQC spectra decoupled in all frequency dimensions. J Magn Reson 199:192–198
Salvador P, Tsai IH, Dannenberg JJ (2011) J-coupling constants for a trialanine peptide as a function of dihedral angles calculated by density functional theory over the full Ramachandran space. Phys Chem Chem Phys 13:17484–17493
Schanda P (2009) Fast-pulsing longitudinal relaxation optimized techniques: enriching the toolbox of fast biomolecular NMR spectroscopy. Prog Nucl Magn Reson Spectrosc 55:238–265
Serber Z, Richter C, Moskau D, Böhlen JM, Gerfin T, Marek D, Häberli M, Baselgia L, Laukien F, Stern AS, Hoch JC, Dötsch V (2000) New carbon-detected protein NMR experiments using cryoprobes. J Am Chem Soc 122:3554–3555
Sinnaeve D, Foroozandeh M, Nilsson M, Morris GA (2016) A general method for extracting individual coupling constants from crowded 1H NMR spectra. Angew Chem Int Ed 55:1090–1093
Smith MA, Hu H, Shaka AJ (2001) Improved broadband inversion performance for NMR in liquids. J Magn Reson 151:269–283
Solyom Z, Schwarten M, Geist L, Konrat R, Willbold D, Brutscher B (2013) BEST-TROSY experiments for time-efficient sequential resonance assignment of large disordered proteins. J Biomol NMR 55:311–321
Uhrín D, Liptaj T, Kövér KE (1993) Modified BIRD pulses and design of heteronuclear pulse sequences. J Magn Reson A 101:41–46
Wang AC, Grzesiek S, Tschudin R, Lodi PJ, Bax A (1995) Sequential backbone assignment of isotopically enriched proteins in D2O by deuterium-decoupled HA (CA)N and HA (CACO)N. J Biomol NMR 5:376–382
Weber PL, Brown SC, Mueller L (1987) Sequential 1H NMR assignments and secondary structure identification of human ubiquitin. Biochemistry 26:7282–7290
Ying J, Roche J, Bax A (2014) Homonuclear decoupling for enhancing resolution and sensitivity in NOE and RDC measurements of peptides and proteins. J Magn Reson 241:97–102
Zangger K (2015) Pure shift NMR. Prog Nucl Magn Reson Spectrosc 86–87:1–20
Zangger K, Sterk H (1997) Homonuclear broadband-decoupled NMR spectra. J Magn Reson 124:486–489
Acknowledgements
B.L. gratefully acknowledges funding by the DFG (LU835/13-1) and the HGF program Information (43.35.02). B.A. thanks funding from NKFI (National Research, Development and Innovation) Grants K124900, K137940. J.D.H. thanks C. Muhle-Goll for a helpful discussion.
Funding
Open Access funding enabled and organized by Projekt DEAL.
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Haller, J.D., Bodor, A. & Luy, B. Pure shift amide detection in conventional and TROSY-type experiments of 13C,15N-labeled proteins. J Biomol NMR 76, 213–221 (2022). https://doi.org/10.1007/s10858-022-00406-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10858-022-00406-z