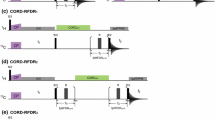
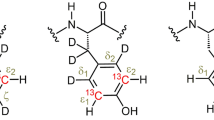
Abstract
Nuclear magnetic resonance spectroscopy (NMR) can provide a great deal of information about structure and dynamics of biomolecules. The quality of an NMR structure strongly depends on the number of experimental observables and on their accurate conversion into geometric restraints. When distance restraints are derived from nuclear Overhauser effect spectroscopy (NOESY), stereo-specific assignments of prochiral atoms can contribute significantly to the accuracy of NMR structures of proteins and nucleic acids. Here we introduce a series of NOESY-based pulse sequences that can assist in the assignment of chiral CHD methylene protons in random fractionally deuterated proteins. Partial deuteration suppresses spin-diffusion between the two protons of CH2 groups that normally impedes the distinction of cross-relaxation networks for these two protons in NOESY spectra. Three and four-dimensional spectra allow one to distinguish cross-relaxation pathways involving either of the two methylene protons so that one can obtain stereospecific assignments. In addition, the analysis provides a large number of stereospecific distance restraints. Non-uniform sampling was used to ensure optimal signal resolution in 4D spectra and reduce ambiguities of the assignments. Automatic assignment procedures were modified for efficient and accurate stereospecific assignments during automated structure calculations based on 3D spectra. The protocol was applied to calcium-loaded calbindin D9k. A large number of stereospecific assignments lead to a significant improvement of the accuracy of the structure.





Similar content being viewed by others
References
Atreya HS, Chary KVR (2001) Selective ‘unlabeling’ of amino acids in fractionally C-13 labeled proteins: an approach for stereospecific NMR assignments of CH3 groups in Val and Leu residues. J Biomol NMR 19:267–272
Bohlen JM, Bodenhausen G (1993) Experimental aspects of chirp NMR-spectroscopy. J Magn Reson A 102:293–301
Cai ML, Liu JH, Gong YX, Krishnamoorthi R (1995) A practical method for stereospecific assignments of gamma-methylene and delta-methylene hydrogens via estimation of vicinal H-1–H-1 coupling-constants. J Magn Reson, Ser B 107:172–178. doi:10.1006/jmrb.1995.1074
Carlomagno T, Peti W, Griesinger C (2000) A new method for the simultaneous measurement of magnitude and sign of D-1(CH) and D-1(HH) dipolar couplings in methylene groups. J Biomol NMR 17:99–109. doi:10.1023/a:1008346902500
Castellani F, van Rossum B, Diehl A, Schubert M, Rehbein K, Oschkinat H (2002) Structure of a protein determined by solid-state magic-angle-spinning NMR spectroscopy. Nature 420:98–102. doi:10.1038/nature01070
Clore GM, Appella E, Yamada M, Matsushima K, Gronenborn AM (1990) 3-dimensional structure of interleukin-8 in solution. Biochemistry 29:1689–1696. doi:10.1021/bi00459a004
Clore GM, Bax A, Gronenborn AM (1991a) Stereospecific assignment of β-methylene protons in larger proteins using 3D 15 N-separated Hartmann–Hahn and 13C-separated rotating frame Overhauser spectroscopy. J Biomol NMR 1:13–22. doi:10.1007/bf01874566
Clore GM, Wingfield PT, Gronenborn AM (1991b) High-resolution 3-dimensional structure of interleukin-1-beta in solution by 3-dimensional and 4-dimensional nuclear-magnetic-resonance spectroscopy. Biochemistry 30:2315–2323. doi:10.1021/bi00223a005
Curley RW, Panigot MJ, Hansen AP, Fesik SW (1994) Stereospecific assignments of glycine in proteins by stereospecific deuteration and N-15 labeling. J Biomol NMR 4:335–340
Driscoll PC, Gronenborn AM, Clore GM (1989) The influence of stereospecific assignments on the determination of 3-dimensional structures of proteins by nuclear magnetic-resonance spectroscopy—application to the sea-anemone protein BDS-I. FEBS Lett 243:223–233
Emerson SD, Montelione GT (1992) Accurate measurements of proton scalar coupling-constants using C-13 isotropic mixing spectroscopy. J Am Chem Soc 114:354–356. doi:10.1021/ja00027a052
Emsley L, Bodenhausen G (1990a) Gaussian pulse cascades-new analytical functions for rectangular selective inversion and in-phase excitation in NMR. Chem Phys Lett 165:469–476
Emsley L, Bodenhausen G (1990b) Phase-shifts induced by transient Bloch–Siegert effects in NMR. Chem Phys Lett 168:297–303
Folmer RHA, Hilbers CW, Konings RNH, Nilges M (1997) Floating stereospecific assignment revisited: application to an 18 kDa protein and comparison with J-coupling data. J Biomol NMR 9:245–258. doi:10.1023/a:1018670623695
Gardner KH, Kay LE (1998) The use of H-2, C-13, N-15 multidimensional NMR to study the structure and dynamics of proteins. Annu Rev Biopys Biomol Struct 27:357–406
Geen H, Freeman R (1991) Band-selective radiofrequency pulses. J Magn Reson 93:93–141
Grzesiek S, Ikura M, Clore GM, Gronenborn AM, Bax A (1992) A 3D triple-resonance NMR technique for qualitative measurement of carbonyl-H-beta J couplings in isotopically enriched proteins. J Magn Reson 96:215–221
Grzesiek S, Anglister J, Ren H, Bax A (1993) C-13 line narrowing by H-2 decoupling in H-2/C-13/N-15-enriched proteins—application to triple-resonance 4D J-connectivity of sequential amides. J Am Chem Soc 115:4369–4370. doi:10.1021/ja00063a068
Guntert P, Braun W, Billeter M, Wuthrich K (1989) Automated stereospecific H-1-NMR assignments and their impact on the precision of protein-structure determinations in solution. J Am Chem Soc 111:3997–4004
Güntert P, Billeter M, Ohlenschläger O, Brown L, Wüthrich K (1998) Conformational analysis of protein and nucleic acid fragments with the new grid search algorithm FOUND. J Biomol NMR 12:543–548. doi:10.1023/a:1008391403193
Hahnke MJ, Richter C, Heinicke F, Schwalbe H (2010) The HN(COCA)HAHB NMR experiment for the stereospecific assignment of H(beta)-protons in non-native states of proteins. J Am Chem Soc 132:918–919. doi:10.1021/ja909239w
Herrmann T, Guntert P, Wuthrich K (2002a) Protein NMR structure determination with automated NOE assignment using the new software CANDID and the torsion angle dynamics algorithm DYANA. J Mol Biol 319:209–227. doi:10.1016/s0022-2836(02)00241-3
Herrmann T, Guntert P, Wuthrich K (2002b) Protein NMR structure determination with automated NOE-identification in the NOESY spectra using the new software ATNOS. J Biomol NMR 24:171–189. doi:10.1023/a:1021614115432
Kainosho M, Torizawa T, Iwashita Y, Terauchi T, Ono AM, Guntert P (2006) Optimal isotope labelling for NMR protein structure determinations. Nature 440:52–57. doi:10.1038/nature04525
Kazimierczuk K, Zawadzka A, Koźmiński W, Zhukov I (2008) Determination of spin–spin couplings from ultrahigh resolution 3D NMR spectra obtained by optimized random sampling and multidimensional Fourier transformation. J Am Chem Soc 130:5404–5405. doi:10.1021/ja800622p
Kazimierczuk K, Stanek J, Zawadzka-Kazimierczuk A, Kozminski W (2010) Random sampling in multidimensional NMR spectroscopy. Prog Nucl Magn Reson Spectrosc 57:420–434. doi:10.1016/j.pnmrs.2010.07.002
Kordel J, Skelton NJ, Akke M, Chazin WJ (1993) High-resolution solution structure of calcium-loaded calbindin-D(9 k). J Mol Biol 231:711–734
Kordel J, Pearlman DA, Chazin WJ (1997) Protein solution structure calculations in solution: solvated molecular dynamics refinement of calbindin D-9k. J Biomol NMR 10:231–243
Kumar A, Ernst RR, Wüthrich K (1980) A two-dimensional nuclear Overhauser enhancement (2D NOE) experiment for the elucidation of complete proton–proton cross-relaxation networks in biological macromolecules. Biochem Biophys Res Commun 95:1–6
Kumar A, Grace RCR, Madhu PK (2000) Cross-correlations in NMR. Prog NMR Spectrosc 37:191–319
Kupce E, Freeman R (1995) Adiabatic pulses for wideband inversion and broadband decoupling. J Magn Reson Ser A 115:273–276
Kushlan DM, Lemaster DM (1993) Resolution and sensitivity enhancement of heteronuclear correlation for methylene resonances via H-2-enrichment and decoupling. J Biomol NMR 3:701–708
Leiting B, Marsilio F, O’Connell JF (1998) Predictable deuteration of recombinant proteins expressed in Escherichia coli. Anal Biochem 265:351–355. doi:10.1006/abio.1998.2904
Lemaster DM (1987) Chiral-beta and random fractional deuteration for the determination of protein side-chain conformation by NMR. FEBS Lett 223:191–196. doi:10.1016/0014-5793(87)80534-3
Lemaster DM (1990) Deuterium labeling in NMR structural-analysis of larger proteins. Q Rev Biophys 23:133–174
Lohr F, Schmidt JM, Ruterjans H (1999) Simultaneous measurement of (3)J(HN, H alpha) and (3)J(H alpha, H beta) coupling constants in C-13, N-15-labeled proteins. J Am Chem Soc 121:11821–11826. doi:10.1021/ja991356h
Luthy J, Retey J, Arigoni D (1969) Preparation and detection of chiral methyl groups. Nature 221:1213. doi:10.1038/2211213a0
Marino JP, Diener JL, Moore PB, Griesinger C (1997) Multiple-quantum coherence dramatically enhances the sensitivity of CH and CH2 correlations in uniformly C-13-labeled RNA. J Am Chem Soc 119:7361–7366. doi:10.1021/ja964379u
Miclet E, Williams DC, Clore GM, Bryce DL, Boisbouvier J, Bax A (2004) Relaxation-optimized NMR spectroscopy of methylene groups in proteins and nucleic acids. J Am Chem Soc 126:10560–10570. doi:10.1021/ja047904v
Mueller L (1987) PE-COSY, a simple alternative to E-COSY. J Magn Reson 72:191–196. doi:10.1016/0022-2364(87)90188-0
Muhandiram DR, Yamazaki T, Sykes BD, Kay LE (1995) Measurement of 2H T1rho relaxation times in uniformly 13C-labeled and fractionally 2H-labeled proteins in solution. J Am Chem Soc 117:11536–11544
Muller L (1979) Sensitivity enhanced detection of weak nuclei using heteronuclear multiple quantum coherence. J Am Chem Soc 101:4481–4484
Neri D, Szyperski T, Otting G, Senn H, Wuthrich K (1989) Stereospecific nuclear magnetic-resonance assignments of the methyl-groups of valine and leucine in the DNA-binding domain of the 434-repressor by biosynthetically directed fractional C-13 labeling. Biochemistry 28:7510–7516
Neuhaus D, Williamson MP (2000) The nuclear Overhauser effect in structural and conformational analysis, 2nd edn. Wiley, New York
Nietlispach D et al (1996) An approach to the structure determination of larger proteins using triple resonance NMR experiments in conjunction with random fractional deuteration. J Am Chem Soc 118:407–415. doi:10.1021/ja952207b
Oktaviani NA, Otten R, Dijkstra K, Scheek RM, Thulin E, Akke M, Mulder FAA (2011) 100% complete assignment of non-labile (1)H, (13)C, and (15)N signals for calcium-loaded calbindin D(9 k) P43G. Biomol NMR Assign 5:79–84. doi:10.1007/s12104-010-9272-3
Orts J, Vögeli B, Riek R, Güntert P (2013) Stereospecific assignments in proteins using exact NOEs. J Biomol NMR 57:211–218. doi:10.1007/s10858-013-9780-4
Ostler G et al (1993) Stereospecific assignments of the leucine methyl resonances in the H-1-NMR spectrum of lactobacillus-casei dihydrofolate-reductase. FEBS Lett 318:177–180
Palmer AG, Cavanagh J, Wright PE, Rance M (1991) Sensitivity improvement in proton-detected 2-dimensional heteronuclear correlation NMR-spectroscopy. J Magn Reson 93:151–170. doi:10.1016/0022-2364(91)90036-s
Paquin R, Ferrage F, Mulder FAA, Akke M, Bodenhausen G (2008) Multiple-timescale dynamics of side-chain carboxyl and carbonyl groups in proteins by 13C nuclear spin relaxation. J Am Chem Soc 130:15805
Piotto M, Saudek V, Sklenar V (1992) Gradient-tailored excitation for single-quantum NMR spectroscopy of aqueous solutions. J Biomol NMR 2:661–665. doi:10.1007/BF02192855
Plevin MJ, Hamelin O, Boisbouvier J, Gans P (2011) A simple biosynthetic method for stereospecific resonance assignment of prochiral methyl groups in proteins. J Biomol NMR 49:61–67. doi:10.1007/s10858-010-9463-3
Pristovsek P, Franzoni L (2006) Stereospecific assignments of protein NMR resonances based on the tertiary structure and 2D/3D NOE data. J Comput Chem 27:791–797. doi:10.1002/jcc.20389
Shaka AJ, Keeler J, Frenkiel T, Freeman R (1983) An improved sequence for broad-band decoupling—WALTZ-16. J Magn Reson 52:335–338
Shaka AJ, Barker PB, Freeman R (1985) Computer-optimized decoupling scheme for wideband applications and low-level operation. J Magn Reson 64:547–552
Shaka AJ, Lee CJ, Pines A (1988) Iterative schemes for bilinear operators—application to spin decoupling. J Magn Reson 77:274–293
Stanek J, Augustyniak R, Kozminski W (2012) Suppression of sampling artefacts in high-resolution four-dimensional NMR spectra using signal separation algorithm. J Magn Reson 214:91–102. doi:10.1016/j.jmr.2011.10.009
States DJ, Haberkorn RA, Ruben DJ (1982) A two-dimensional nuclear Overhauser experiment with pure absorption phase in 4 quadrants. J Magn Reson 48:286–292
Svensson LA, Thulin E, Forsén S (1992) Proline cis-trans isomers in calbindin D9k observed by X-ray crystallography. J Mol Biol 223:601–606
Takeda M, Terauchi T, Kainosho M (2012) Conformational analysis by quantitative NOE measurements of the beta-proton pairs across individual disulfide bonds in proteins. J Biomol NMR 52:127–139. doi:10.1007/s10858-011-9587-0
Vallurupalli P, Hansen DF, Lundstrom P, Kay LE (2009) CPMG relaxation dispersion NMR experiments measuring glycine H-1(alpha) and C-13(alpha) chemical shifts in the ‘invisible’ excited states of proteins. J Biomol NMR 45:45–55. doi:10.1007/s10858-009-9310-6
Vögeli B, Segawa TF, Leitz D, Sobol A, Choutko A, Trzesniak D, van Gunsteren W, Riek R (2009) Exact distances and internal dynamics of perdeuterated ubiquitin from NOE buildups. J Am Chem Soc 131:17215–17225. doi:10.1021/ja905366h
Vuister GW, Bax A (1992) Resolution enhancement and spectral editing of uniformly C-13-enriched proteins by homonuclear broad-band C-13 decoupling. J Magn Reson 98:428–435. doi:10.1016/0022-2364(92)90144-v
Wagner G, Braun W, Havel TF, Schaumann T, Go N, Wuthrich K (1987) Protein structures in solution by nuclear-magnetic-resonance and distance geometry—the polypeptide fold of the basic pancreatic trypsin-inhibitor determined using 2 different algorithms, DISGEO and DISMAN. J Mol Biol 196:611–639. doi:10.1016/0022-2836(87)90037-4
Yamazaki T, Yoshida M, Nagayama K (1993) Complete assignments of magnetic resonances of ribonuclease-H from Escherichia-coli by double-resonance and triple-resonance 2D and 3D NMR spectroscopies. Biochemistry 32:5656–5669. doi:10.1021/bi00072a023
Acknowledgments
We thank Mikael Akke (Lund University) for a sample of partially deuterated calbindin D9k, as well as Dominique Frueh (Johns Hopkins University) and Lewis Kay (University of Toronto) for fruitful suggestions. This research was supported by the Polish budget funds for science in 2013–2014 (Project IP2012 057872 awarded to J.S.) Access to the Research Infrastructure at University of Warsaw was financed by the European Commission’s FP7 (Contract 228461, EAST-NMR). Financial support of the Bio-NMR Project No. 261863 is gratefully acknowledged.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Augustyniak, R., Stanek, J., Colaux, H. et al. Nuclear overhauser spectroscopy of chiral CHD methylene groups. J Biomol NMR 64, 27–37 (2016). https://doi.org/10.1007/s10858-015-0002-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10858-015-0002-0