Abstract
Purpose
With advances in immunology, increasing evidence suggests that immunity is involved in premature ovarian insufficiency (POI) pathogenesis. This study investigated the roles of immune checkpoint genes and immune cell infiltration in POI pathogenesis and development.
Methods
The GSE39501 dataset and immune checkpoint genes were obtained from the Gene Expression Omnibus database and related literature. The two datasets were intersected to obtain immune checkpoint-related differentially expressed genes (ICRDEGs), which were analyzed using Gene Ontology and Kyoto Encyclopedia of Gene and Genomes enrichment analysis, weighted correlation network analysis, protein–protein interaction and related microRNAs, transcription factors, and RNA binding proteins. The immune cell infiltration of ICRDEGs was explored, and receiver operating characteristic curves were used to validate the diagnostic value of ICRDEGs in POI.
Results
We performed ICRDEG functional enrichment analysis and found that these genes were closely related to immune processes, such as T cell activation. Specifically, they are enriched in various biological processes and pathways, such as cell adhesion molecule and T cell receptor signaling pathways. Weighted correlation network analysis identified seven hub genes: Cd200, Cd274, Cd28, neurociliary protein-1, Cd276, Cd40lg, and Cd47. Furthermore, we identified 112 microRNAs, 17 RNA-binding proteins, and 101 transcription factors. Finally, immune infiltration analysis showed a clear positive correlation between hub genes and multiple immune cell types.
Conclusion
Bioinformatic analysis identified seven potential ICRDEGs associated with POI, among which the immune checkpoint molecules CD200 and neurociliary protein-1 may be involved in the pathogenesis of POI.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
POI is defined as amenorrhea with high gonadotropin and low estrogen levels in women younger than 40 years [1]. The cessation of menses at a younger age leads to infertility and decline in psychological well-being and long-term quality of life of these patients. The pathogenesis of POI is diverse, including iatrogenic factors (post-surgery, radiotherapy, or chemotherapy), autoimmune ovarian function impairment, environmental factors (viruses, chemical agents, and radiation), metabolic diseases (diabetes type 1, galactosemia, 17-OH deficiency, and 21-OH deficiency), and unexplained factors [2]. Given the absence of an effective method to restore ovarian function in POI [3], it is necessary to identify markers of POI for earlier identification and intervention.
The immune system is composed of several relevant molecular and chemical systems, their primary function is to protect the body from pathogens. The immune system plays multiple roles in the female reproductive system, mainly in gonadal activity and regulation of the female reproductive tract, and ovarian cycle [4]. Disease may occur if normal physiological interactions between the immune and reproductive systems are dysregulated.
Immune checkpoint molecules exist on the surface of immune cells, and the negative regulation of effector immune cells prevents the body from being attacked by its own antigens [5]. The PD-1 immune checkpoint pathway is involved in follicle formation and ovulation [6, 7]. Little is known about the normal interactions between female germ and immune cells, necessitating this relationship to be characterized.
Based on bioinformatics analysis, this study explored the possible pathophysiological process of ovarian disease from the perspective of immune checkpoint molecules related to POI based on the Weighted Gene Association Network Analysis (WGCNA) and immune infiltration. Further studies on the relation between POI and immune checkpoint genes, as well as immune cell infiltration, could provide new insights into the immune mechanisms associated with POI.
Materials and Methods
Data resource
We used “premature ovarian insufficiency” and “premature ovarian failure” as keyword on the Gene Expression Omnibus(GEO) database, microarray datasets were obtained with the accession no [8, 9]. Dataset GSE39501, obtained from Mus musculus, comprised samples from patients with POI and normal samples, totaling six samples. The data platform file used was the GPL6887 Illumina MouseWG-6 v2.0 expression beadchip. We included the expression profile data of three samples from patients with POI (POI group) and three normal samples (Normal group). The details of the dataset are listed in Table 1.
The published literatures included in PubMed database were searched with the key words “immune checkpoint genes”, and finally we identified 73 immune checkpoint genes for analysis [10] (see Table S1).
Differentially expressed genes (DEGs) associated with POI
We performed differential analysis of dataset GSE39501 using the softwear package in R used for analysis of differential gene expression [11] to obtain DEGs among the Normal and POI groups, and genes with |logFC|> 0 and P-value < 0.05 were selected as DEGs for further study. Genes with logFC > 0 and P-value < 0.05 were DEGs (upregulated genes), and those with logFC < 0 and P-value < 0.05 were DEGs (downregulated genes).
To obtain the immune checkpoint-related DEGs (ICRDEGs) associated with POI, we crossed the DEGs in the GSE39501 dataset with immune checkpoint-related genes and drew a Venn diagram, the results of the differential analysis were presented in a volcano map and heat map using the R package ggplot2.
Functional and pathway enrichment analyses of DEGs
We performed Gene Ontology (GO) [12] and Kyoto Encyclopedia of Genes and Genomes (KEGG) [13] annotation analysis of ICRDEGs using the R package clusterProfiler [14]. Entry screening criteria of P-value < 0.05 and FDR value (q.value) < 0.05 were considered statistically significant, with P-value correction applied using the Benjamini–Hochberg method.
Gene Set Enrichment Analysis (GSEA)
We obtained data from the Molecular Signatures Database “m2.all.v2022.1” gene set, specifically using the “Mm.symbols.gmt” gene set. GSEA [15] was performed with the expressed genes in the GSE39501 dataset, with the screening criteria P-value < 0.05 and FDR value (q.value) < 0.25 indicating significant enrichment.
Gene Set Variation Enrichment Analysis (GSVA)
We obtained the “m2.all.v2022.1” gene set from Molecular Signatures Database. GSVA [16] was used to analyze the GSE39501 dataset for calculating the difference in functional enrichment between the Normal and POI groups.
WGCNA
We performed WGCNA [17] on the GSE39501 dataset using the R package WGCNA. Selected genes with |logFC|> 0 and P-value < 0.05 were inputted in the WGCNA. The minimum number of module genes was set to 100, and the softpower was optimized with a soft threshold of 14. The combined shear height of the modules was set to 0.2, and the minimum distance was set to 0.2 to assess the correlation of the Normal and POI groups with different modules. Genes in each module were recorded, and module eigengenes were considered. After selecting the modules of interest based on the correlation values, we identified all genes within the module as DEGs that are highly associated with POI.
Immune infiltration analysis
In this study, the immune cell infiltration status in the GSE39501 dataset was evaluated using the CIBERSORT (https://cibersort.stanford.edu/) [18] algorithm. Spearman correlation was employed to assess the correlation between various immune cells, and thus, determine the correlation between genes with high correlation to candidate and immune infiltrating cells. The correlation between immune cells and ICRDEGs combined with the gene expression matrix of the POI dataset was then assessed, and the correlation heat map was drawn using the R package pheatmap.
The enrichment scores calculated by the single-sample GSEA (ssGSEA) [19] algorithm in the R package GSVA were used to represent the infiltration level of each immune cell type in each sample, and the correlation between the immune infiltrating cells was determined by Spearman correlation analysis.
For Spearman correlation analysis based on MCPcounter abundance estimates as well as expression of antigen genes, a P-value < 0.05 was considered statistically significant.
Protein–protein interaction (PPI) networks and mRNA-microRNA (miRNA) mRNA-RNA binding protein (RBP) mRNA-transcription factor (TF) interaction networks
In this study, we used the STRING database [20] to screen the ICRDEGs selected from the differential analysis to construct PPI networks associated with DEGs and to visualize the PPI network model using Cytoscape. The closely linked local regions of the PPI network, which may represent molecular complexes, have specific biological functions.
The ENCORI [21] (https://starbase.sysu.edu.cn/) and miRDB databases [22] were used for miRNA target gene prediction and functional annotation. We used the ENCORI and miRDB databases to predict the miRNAs interacting with hub genes (mRNA) and then utilized the intersection part of the results of the two databases to draw the mRNA-miRNA interaction network.
We searched for TFs bound to hub genes (mRNA) using the CHIPBase database [23] (version 3.0; https://rna.sysu.edu.cn/chipbase/).
Correlation analysis and receiver operating characteristic curve analysis of DEGs
The pROC package was used to plot the receiver operating characteristic curves of hub genes in the Normal and POI groups in the two datasets, and the area under the curve was calculated to evaluate the diagnostic value of hub gene expression for POI.
Statistical analysis
All data processing and analyses in this study were performed using R software (Version 4.1.2). For the comparison of two continuous variables, the statistical significance of normally distributed variables was estimated using the independent Student’s t-test, and differences between non-normally distributed variables were analyzed using the Mann–Whitney U test (i.e., Wilcoxon rank sum test). Chi-square or Fisher's exact tests were used to compare and analyze the statistical significance between two groups of categorical variables. If not explicitly specified, the results were calculated as correlation coefficients between different molecules by Spearman correlation analysis, and all P-values were two-sided, with P < 0.05 considered statistically significant.
Results
Technical roadmap
Data standardization
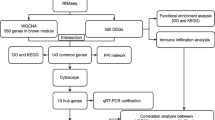
This study mainly explored the biological characteristics of POI using bioinformatics methods, and a flow chart of the overall analysis is shown in Fig. 1. In the GSE39501 dataset, the samples were divided into the POI and Normal groups. Subsequently, the GSE39501 dataset (Fig. 2A–B) was standardized, data cleaning operations, such as annotated probes, were performed, and the data distribution before and after standardization was boxed. After normalization, the expression trends converged across different samples in the dataset.
Technical Roadmap. POI: Premature ovarian insufficiency. GSEA: Gene Set Enrichment Analysis. GSVA: Gene Set Variation Analysis. ssGSEA: single-sample Gene Set Enrichment Analysis. GO: Gene Ontology. KEGG: Kyoto Encyclopedia of Genes and Genomes. PPI network: Protein–Protein Interaction network. ROC: Receiver Operating Characteristic PCA: Principal Component Analysis. ICRDEGs: immune checkpoint-related differentially expressed genes. DEGs: Differentially Expressed Senes
Boxplot before and after correction of the GSE39501 dataset for POI. A. Box plot of gene expression distribution between samples in the GSE39501 dataset before correction. B. Box plot of gene expression distribution among samples for the corrected GSE39501 dataset. Yellow represents the normal group and purple represents the POI sample group. POI: Premature ovarian insufficiency
Analysis of DEGs associated with POI
There were 9209 DEGs in total, including 4309 genes being up-regulated, and 4900 genes being down-regulated. We visualized the results with a volcano map based on the differential analysis of this dataset (Fig. 3A).
Differential gene analysis of the premature ovarian insufficiency dataset. A. Volcano plots of differential genes in the GSE39501 dataset. B. Venn diagram of the DEGs and ICRGs in the GSE39501 dataset. C. Heat map of differentially expressed genes in the GSE39501 dataset. DEGs: differentially expressed genes. ICRGs: immune checkpoint-related genes
To obtain ICRDEGs, we took the intersection of DEGs and immune checkpoint-related genes with |logFC|> 0 and P-values < 0.05 obtained in the GSE39501 dataset. This process yielded 14 ICRDEGs. The results were visualized using a Venn diagram (Fig. 3B).
We also analyzed the differences in expression between POI and Normal groups in the GSE39501 dataset (Fig. 3C) and used the R package pheatmap to generate a heat map illustrating the expression patterns of the 14 identified ICRDEGs (Btla, Btnl2, Cd160, Cd200, Cd274, Cd276, Cd28, Cd40lg, Cd44, Cd47, G0s2, Klrd1, neurociliary protein-1 [Nrp1], and Pdcd1). The results showed that 14 DEGs clustered significantly in the two groups within the GSE39501 dataset.
Functional and pathway enrichment analyses of ICRDEGs
To analyze the biological processes associated with the 14 ICRDEGs along with their molecular function, cellular component, biological pathways, and relation with POI, we first performed GO (Table 2) and KEGG (Table 3) enrichment analysis for ICRDEGs. The screening criterion for enrichment entries was defined as P-value < 0.05, with an associated FDR value (q.value) < 0.05, considered statistically significant. The results of GO functional enrichment analysis and KEGG enrichment analysis are displayed as a bubble map (Fig. 4A–B) and a ring network map (Fig. 4C–D). Subsequently, we showed the results of GO function enrichment analysis of ICRDEGs combined with logFC (Fig. 4E–F).
Functional enrichment analysis (GO) and pathway enrichment (KEGG) analysis of ICRDEGs. A–B. Bubble diagram display of GO function enrichment analysis (A) and KEGG pathway enrichment analysis (B) of ICRDEGs. C–D. Ring network diagram display of GO function enrichment analysis (C) and KEGG pathway enrichment analysis (D) of ICRDEGs. E–F. GO function enrichment analysis (E) and KEGG pathway enrichment analysis (F) of ICRDEGs. ICRDEGs: immune checkpoint-related differentially expressed genes. GO: Gene Ontology. BP: Biological Orocess. CC: Cellular Component. MF: Molecular Function. KEGG: Kyoto Encyclopedia of Genes and Genomes. The screening criteria for the GO and KEGG enrichment entries were P-value < 0.05 and FDR value (q.value) < 0.05
The results of these analyses showed that the genes were mainly enriched in the regulation immunity and inflammation biological process(GO:0032088). For the cellular component, the gene were mainly enriched the carriers of biomarkers in extracellular spaces(GO:0070062). Finally, regarding molecular function, the genes were mainly enriched in the receptor function(GO:0017154). Furthermore, KEGG enrichment analysis revealed that the ICRDEGs were mainly associated with pathways, such as cell adhesion molecules (mmu04514), T cell receptor signaling pathway (mmu04660), allograft rejection (mmu05330), graft-versus-host disease (mmu05332), autoimmune thyroid disease (mmu05320), extracellular matrix-receptor interaction (mmu04512), viral myocarditis (mmu05416), and primary immunodeficiency (mmu05340).
We used the R package Pathview (Fig. 5) to illustrate the results for cell adhesion molecules (Fig. 5A), T cell receptor signaling pathway (Fig. 5B), allograft rejection (Fig. 5C), primary immunodeficiency (Fig. 5D), and four KEGG pathways in the KEGG enrichment analysis.
Pathway enrichment (KEGG) analysis of ICRDEGs. A–D Pathway enrichment (KEGG) enrichment analyses results for ICRDEGs. KEGG pathway cell adhesion molecules (A), T cell receptor signaling pathway (B), allograft rejection (C), and primary immunodeficiency (D). ICRDEGs: immune checkpoint-related differentially expressed genes. KEGG: Kyoto Encyclopedia of Genes and Genomes
GSEA and GSVA enrichment analysis of the POI dataset
To determine the effect of gene expression levels on POI, we analyzed the links between gene expression in the Normal and POI groups and biological processes, affected cellular components, and molecular functions using GSEA enrichment analysis. All genes in the GSE39501 dataset were significantly enriched in the phosphatidylinositol 3-kinase (PI3K)-AKT pathway (Fig. 6B), MAPK pathway (Fig. 6C), TGFbeta pathway (Fig. 6D), WNT pathway (Fig. 6E), NOTCH pathway (Fig. 6F), HEDGEHOG pathway (Fig. 6G), TP53 pathway (Fig. 6H), and other pathways (Table 4).
GSEA and GSVA enrichment analysis for the GSE39501 Dataset. A. GSEA enrichment analysis of the GSE39501 dataset. B–H. Differential genes in the GSE39501 dataset were significantly enriched in the PI3K-AKT pathway (B), MAPK pathway (C), TGFBETA pathway (D), WNT pathway (E), NOTCH pathway (F), HEDGEHOG pathway (G), and TP53 pathway (H). I. GSVA analysis of the GSE39501 dataset. Yellow represents the Normal group and purple represents the POI group. POI: premature ovarian insufficiency. GSEA: Gene Set Enrichment Analysis. GSVA: Gene Set Variation Analysis. The screening criteria for determining significant enrichment in GSEA and GSVA enrichment analyses were P-value < 0.05 and FDR value (q.value) < 0.25
To explore the differences in the GSE39501 dataset between the POI samples and the corresponding normal samples, we performed GSVA enrichment analysis (Fig. 6I) and assessed the differences in functional enrichment. The results showed that the IL-2 pathway isogene set showed differences between the POI and Normal groups (Table 5).
WGCNA analysis to identify the co-expression module in the GSE39501 dataset
We performed a WGCNA of all |logFC|-expressed genes (DEGs) in the GSE39501 dataset to screen for co-expression modules. In the course of the WGCNA, we first clustered the POI and Normal groups in GSE39501 using cluster tree and annotated the grouping information (without setting cut height). Subsequently, we set a screening criterion of 0.9 to determine the optimal number of modules. DEGs were clustered in 14 modules: MEcyan, MEdarkgrey, MEdarkred, MEorange, MEred, MEdarkorange, MEmidnightblue, MEpurple, MEdarkturquoise, MEgrey60, MElightyellow, MElightcyan, MEblue, and MEmagenta (Fig. 7A). Subsequently, we performed another round of clustering of the DEGs, visualizing the relation between the genes and their corresponding new modules. Finally, according to the expression pattern of the module genes and the different grouping information of the GSE39501 dataset, we obtained 14 modules (MEcyan, MEdarkgrey, MEdarkred, MEorange, MEred, MEdarkorange, MEmidnightblue, MEpurple, MEdarkturquoise, MEgrey60, MElightyellow, MElightcyan, MEblue, and MEmagenta). The correlation between the two groups in GSE39501 is depicted in (Fig. 7B). Subsequently, we applied a module merge shear height of 0.2 to shear and combine modules with a merge shear height below 0.2 (Fig. 7C).
WGCNA analysis identifies the co-expression module in the GSE39501 dataset. A. Sample module screening threshold in the GSE39501 dataset. B. Correlation analysis between clustering modules of DEGs and different groups in the GSE39501 dataset. C. Results of DEGs in the GSE39501 dataset. D-I. DEGs in the GSE39501 dataset shown in Venn diagrams. MEblue (D), MEdarkgrey (E), MEdarkred (F), MEmidnightblue (G), MEorange (H), and MEred (I). J. Chromosomal localization map of hub genes. K. Visual presentation of hub genes Friends analysis. WGCNA: Weighted Gene Association Network Analysis. POI: Premature Ovarian Insufficiency. DEGs: Differentially Expressed Genes. ICRDEGs: immune checkpoint-related differentially expressed genes
First, we conducted a subsequent analysis of DEGs in the 14 modules (MEblue, MEdarkgrey, MEdarkred, MEmidnightblue, MEred, and MEorange) (P < 0.05, absolute correlation value 0.5). Second, we crossed the ICRDEGs in the GSE39501 dataset and the DEGs included in the six modules and generated a Venn diagram (Fig. 7D–I) to obtain the modules. As shown in Fig. 7, we obtained seven ICRDEGs (Cd200, Cd274, Cd28, Nrp1, Cd276, Cd40lg, and Cd47) as hub genes for our subsequent study.
We annotated the human chromosome locations for the seven hub genes and visualized them in a circle (Fig. 7J). The chromosome locations are as follows: CD28 on chromosome 2, CD47 and CD200 on chromosome 3, CD274 on chromosome 6, NRP1 on chromosome 10, CD276 on chromosome 15, and CD40LG on chromosome X.
We performed a functional similarity analysis of the seven hub genes by calculating the GO terms using the R package GOSemSim package. The analysis included the identification of GO Term Set (sets of GO terms), measuring the Semantic similarity between the gene products and gene cluster. Subsequently, the results of the functional similarity analysis between the seven hub genes were visualized through a boxplot (Fig. 7K). The results showed that Cd274 has the highest functional similarity with the other hub genes.
Expression and correlation analyses of the hub genes in the dataset
To further explore the differences in expression of the hub genes in the POI dataset, we selected the seven hub genes screened by constructing the WGCNA network (Cd200, Cd274, Cd28, Nrp1, Cd276, Cd40lg, and Cd47) and further analyzed the correlation between the expression level in the GSE39501 dataset (Fig. 8A) and the Normal and POI groups. The results showed that the expression of hub gene Cd200 in the GSE39501 dataset was statistically significant (P < 0.001). The expression levels of hub genes Cd274, Cd276, Cd40lg, Cd47, and Nrp1 were significantly different between the Normal and POI groups (P < 0.01). Additionally, the expression levels of hub gene Cd28 showed statistical significance in the Normal and POI groups (P < 0.05).
Differential expression and correlation analyses of the hub genes in the GSE39501 dataset. A. Comparative analysis of the expression levels of the hub genes in the GSE39501 dataset. B–H Scatter plots displaying correlation analysis results (CorResults) of the hub genes in the GSE39501 dataset. The symbol * indicates P < 0.05, which represents some statistical significance. The symbol * * indicates P < 0.01, representing high statistical significance. The symbol * * * indicates P < 0.001 and represents very high statistical significance. ICRDEGs: immune checkpoint-related differentially expressed genes. POI: premature ovarian insufficiency
To further assess the correlation between the seven hub genes, we generated a heat map of the correlation between genes (Fig. 8B–H) and noted a statistically significant difference between Cd47 and Cd40lg (P = 0.033). The correlation between Nrp1 and Cd200 showed a highly significant difference (P = 0.003).
To explore the differences in the expression of the seven hub genes in the POI dataset, we verified the differential expression of the genes among the Normal and POI groups, and the results are shown in Supplementary File Fig. S1.
PPI and mRNA-miRNA mRNA-TF mRNA-RBP interaction networks
We used the STRING database for seven ICRDEGs (Cd200, Cd274, Cd28, Nrp1, Cd276, Cd40lg, and Cd47) for the PPI analysis, taking the minimum intercorrelation coefficient greater than 0.400 (minimum required interaction score: low confidence [0.150]) as the standard. The resulting PPI network was visualized and mapped using Cytoscape software (Fig. 9A).
Protein–protein interaction (PPI) and mRNA-miRNA, mRNA-TF, mRNA-RBP interaction networks. A. The PPI network of the ICRDEGs. B. The hub genes-miRNA interaction network. C. The hub genes-transcription factor interaction network. D. The hub genes-RBP interaction network. Yellow ellipse depicts mRNA. The blue ellipse depicts miRNA. The pink ellipse depict transcription factors. The green ellipse depicts the RBP. POI: premature ovarian insufficiency
We used the mRNA-miRNA data from the ENCORI and miRDB databases with seven ICRDEGs (Cd200, Cd274, Cd28, Nrp1, Cd276, Cd40lg, and Cd47) for prediction of the interacting miRNAs. Subsequently, the intersection of the two database results was visualized by drawing the mRNA-miRNA interaction network using Cytoscape software (Fig. 9B). Our mRNA-miRNA interaction network is composed of seven hub genes (mRNA) (Cd200, Cd274, Cd28, Nrp1, Cd276, Cd40lg, and Cd47), and a total of 112 miRNA molecules, resulting in 121 mRNA-miRNA interaction pairs (Table S2).
We searched for the TFs bound to the hub genes using the CHIPBase database (version 3.0). The mRNA-TF interaction network was then visualized using Cytoscape software (Fig. 9C), resulting in interaction data for six hub genes (mRNA) (Cd47, Cd200, Cd274, Cd276, Nrp1, and Cd40lg) and 101 TFs, forming 124 mRNA-TF interaction pairs (Table S3).
We used the mRNA-RBP data from the ENCORI database to predict interactions with seven ICRDEGs (Cd200, Cd274, Cd28, Nrp1, Cd276, Cd40lg, and Cd47). The database results were then visualized as an mRNA-RBP interaction network using Cytoscape software (Fig. 9D). Our mRNA-RBP interaction network is composed of six hub genes (mRNA) (Cd200, Cd274, Cd276, Cd28, Cd47, and Nrp1) and 17 RBP molecules, forming a total of 42 mRNA-RBP interaction pairs (Table S4).
Immune infiltration analysis of the POI dataset
We used the CIBERSORT algorithm to calculate the correlation between the expression profile data of the Normal and POI groups from 22 immune cells in the GSE39501 dataset. Based on the results of the immune infiltration analysis, we plotted the infiltration of 22 immune cells in each sample of the GSE39501 dataset (Fig. 10A) using bar charts.
Analysis of immune infiltration in the GSE39501 dataset (CIBERSORT). A. Bar graph of immune infiltration results of 22 immune cells. B–C. Heat map of the correlation analysis of ICRDEGs and immune cell expression. The symbol * indicates P < 0.05, which represents statistical significance. The symbol ** indicates P < 0.01, representing high statistical significance. ICRDEGs: immune checkpoint-related differentially expressed genes. POI: premature ovarian insufficiency. In the heatmap, circles indicate genes with a positive correlation with the infiltration abundance of immune cells. Larger circles indicate stronger correlations. Yellow circles represent a negative correlation between genes and infiltration abundance of immune cells, with larger circles indicating a stronger correlation
We analyzed the differences in gene expression in 22 immune cells among the Normal and POI groups and generated a group comparison diagram. The results are shown in Supplementary File Fig. S1.
We used the ssGSEA algorithm to calculate the correlation between the expression profile data of the Normal and POI groups. Based on the results of the immune infiltration analysis, we identified a correlation between seven hub genes (Cd200, Cd274, Cd28, Nrp1, Cd276, Cd40lg, and Cd47) (Fig. 10B) (P < 0.05). In the GSE39501 dataset, a strong correlation was observed between the abundance of infiltrating immune cells, immature B cells, and CD40LG.
We assessed the correlation between the expression profiles of the Normal and POI groups in the GSE39501 dataset (Fig. 10C) using the MCPCounter algorithm. MCPCounter showed that the NRP1, CD47, CD40LG, CD28, CD276, CD274, and CD200 genes were significantly positively associated with a variety of immune cells.
Discussion
In a previous study, Wagner et al. used single-cell sequencing to demonstrate macrophages and T cells account for 0.5% of the ovarian cortex [24]. These immune cells can be involved in abnormal ovarian functions, such as abnormal ovulation [25], reduced oocyte number [26], destruction of the ovarian periodic angiogenesis system [27], and increased follicular atresia [25]. However, at present, the mechanism of ovarian dysfunction caused by immune-related reasons is not clear, and there is no specific immune diagnostic index for early diagnosis of ovarian failure. Therefore, it is necessary to elucidate the immune mechanisms related to ovarian disease. In this study, we first identified DEGs and immune checkpoint genes associated with POI. We then analyzed their related pathways and biological functions, and used WGCNA to select seven core genes. We finally performed a series of analyses of core genes to analyze and predict their associated potentially pathogenic mechanism.
There are 14 ICRDEGs, according to GO function enrichment analysis, ICRDEGs were mainly involved in the regulation immunity and inflammation biological process. The four pathways enriched in KEGG analysis were all involved in the T cell receptor signaling pathway. T cell-mediated immune responses can be divided into T cell activation, proliferation, and differentiation. T cell activation requires co-stimulation of dual signals, the first signal comes from the antigen and requires the interaction and binding of the major histocompatibility complex-antigen peptide complex on the antigen-presenting cell surface with T cell receptor, which ensures antigen specificity of the immune response. The second signal is a costimulatory molecule, which ensures that the immune response occurs only under the desired conditions. The dual-signal stimulation mode of T cell activation is a fail-safe mechanism, and the second signal ensures the correct timing and site of initiating T cell response. The lack of a second signal can keep autoreactive T cells in a nonresponsive state, which favors autoimmune tolerance and prevents the activation of autoreactive T lymphocytes. CD28 and CTLA-4 are involved in the regulation of T cell activation. Studies have shown that genetic defects in CTLA-4 lead to CD28-mediated severe autoimmunity [28]. Based on our analysis, we can suspect POI-related immune checkpoint genes are involved in abnormal T cell activation, potentially contributing to the occurrence of disease. This insight provides a promising direction for future investigations into the relationship between T cell activation and POI.
According to the GSVA results, all genes in the GSE39501 datset were significantly enriched in the phosphatidylinositol 3-kinase-AKT pathway, MAPK pathway, TGFbeta pathway, WNT pathway, NOTCH pathway, HEDEGHOG pathway and TP53 pathway. Members of the phosphatidylinositol 3-kinase family has important functions in the immune system. PI3K is an important downstream effector of BCR signaling, further activating the expression of other genes involved in B cell proliferation, differentiation, and innate immune [29]. The MAPK pathway is a canonical inflammatory pathway and key regulator of the inflammatory response and innate immunity. The GSVA results showed that the IL-2 pathway isogene set showed differences between the POI and Normal groups. IL-2 is the result of T cell stimulation and further promotes T cell stimulation[30, 31]. The enrichment of the IL-2 pathway suggested an adaptive immune component to POI. These results indicate that the genes and pathways that were significantly different between the disease and normal group samples of the GSE39501 dataset involved both innate and adaptive immunity, as well as inflammatory responses.
The WGCNA-ICRDEG intersection identified seven core genes, Cd200, Cd274, Cd28, Nrp1, Cd276, Cd40lg, and Cd47 as well as 112 miRNAs, 101 TFs, and 17 RBPs regulatory networks for the core genes. Expression and correlation analyses of core genes in the dataset showed that CD200 was differentially expressed in the Normal and POI groups, and the correlation between Nrp1 and CD200 was significant in the POI dataset.
CD200 (OX-2) belongs to the type I membrane glycoprotein and immunoglobulin superfamily [32] and is expressed in epithelial cells, endothelial cells, fibroblasts, lymphocytes, neurons, ovarian granulosa cells, downregulated myeloid cells (neutrophils, macrophages, and dendritic cells). Studies have shown that CD200 regulates immune and inflammatory responses [33] by regulating CD200R to inhibit macrophage activation and transmit immunosuppressive signals to macrophages. Studies have also shown that macrophages are ubiquitous in the female reproductive system, playing physiological roles in the female menstrual cycle and participating in the regulation of the pituitary–gonadal axis [34]. The proportion of macrophage subsets in old mice is significantly increased [26] when compared with that in younger mice. Thus, we need conduct further experimental studies to verify the mechanism by which CD200-mediated POI increases the susceptibility to ovarian immune diseases due to disruptions in macrophages physiological function.
The correlation analyses revealed a significant correlation between Nrp1 and CD200. Nrp1 is a transmembrane glycoprotein with important roles in embryonic tissue development, angiogenesis, and tumor metastasis [35]. Both Nrp1 and CD200 function as negative regulators of the immune response. While their roles in reproductive functions are less explored, the potential involvement of both in the immune pathogenesis of POI requires further research.
The negative regulation of the immune response and physiological function by the core genes previously mentioned serves to prevent immune damage to body tissue and reduce immune activation. Any abnormality in these regulatory mechanisms can lead to unpredictable immune damage. Although the exact causes of POI are still unclear, numerous experiments have demonstrated the involvement of immune system disorders in its pathogenesis [36]. The immune checkpoint molecules discussed previously could serve as focal points for future investigations into immune-related genes.
The results of the immune infiltration analysis showed a strong correlation between the infiltration abundance of immature B cells and CD40LG in the ssGSEA algorithm, indicating that CD40LG may play an important role in regulating or affecting the infiltration abundance of immature B cells. MCPCounter showed that the NRP1, CD47, CD40LG, CD28, CD276, CD274, and CD200 genes were positively associated with a variety of immune cells, implying that the core gene plays an important role in the immune regulation of POI, although the specific regulatory mechanism requires further study.
Conclusions
The results of this study indicate that T cell-mediated immune and inflammatory responses may be involved in the pathogenesis of premature ovarian aging. Furthermore, there is a need for future investigation into the effects of the T cell receptor signaling pathway and immune checkpoint molecules CD200 and NRP1 on the ovarian immune system.
Data availability
Data are available in a public, open access repository. Data are available on reasonable request. All data relevant to the study are included in the article or uploaded as supplemental information.The datasets (GEO data) for this study can be found in the GEO (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE39501).
References
Komorowska B. Autoimmune premature ovarian failure. Prz Menopauzalny. 2016;15(4):210–4. https://doi.org/10.5114/pm.2016.65666.
Menopause Subgroup, Chinese Society of Obstetrics and Gynecology, Chinese Medical Association. Consensus of clinical diagnosis and treatment of premature ovarian insufficiency (2023). Zhonghua Fu Chan Ke Za Zhi. 2023;58(10):721–728. Chinese. https://doi.org/10.3760/cma.j.cn112141-20230316-00122
Dragojević-Dikić S, Marisavljević D, Mitrović A, Dikić S, Jovanović T, Janković-Raznatović S. An immunological insight into premature ovarian failure (POF). Autoimmun Rev. 2010;9(11):771–4. https://doi.org/10.1016/j.autrev.2010.06.008.
Ye H, Li X, Zheng T, et al. The effect of the immune system on ovarian function and features of ovarian germline stem cells. Springerplus. 2016;5:990. https://doi.org/10.1186/s40064-016-2390-3.
Miko E, Meggyes M, Doba K, Barakonyi A, Szereday L. Immune Checkpoint Molecules in Reproductive Immunology. Front Immunol. 2019;10:846. https://doi.org/10.3389/fimmu.2019.00846.
Johnson J, Kim SY, Sam PK, Asokan R, Cari EL, Bales ES, Luu TH, Perez L, Kallen AN, Nel-Themaat L, Polotsky AJ, Post MD, Orlicky DJ, Jordan KR, Bitler BG. Expression and T cell regulatory action of the PD-1 immune checkpoint in the ovary and fallopian tube. Am J Reprod Immunol. 2023;89(3): e13649. https://doi.org/10.1111/aji.13649.
Xu PC, Luan Y, Yu SY, Xu J, Coulter DW, Kim SY. Effects of PD-1 blockade on ovarian follicles in a prepubertal female mouse. J Endocrinol. 2021;252(1):15–30. https://doi.org/10.1530/JOE-21-0209.
Davis S, Meltzer PS. GEOquery: a bridge between the Gene Expression Omnibus (GEO) and BioConductor. Bioinformatics. 2007;23:1846–7. https://doi.org/10.1093/bioinformatics/btm254.
Wurmbach E, et al. Genome-wide molecular profiles of HCV-induced dysplasia and hepatocellular carcinoma. Hepatology (Baltimore Md). 2007;45:938–47. https://doi.org/10.1002/hep.21622.
Xu D, et al. Identification of immune subtypes and prognosis of hepatocellular carcinoma based on immune checkpoint gene expression profile. Biomed Pharmacother Biomed Pharmacother. 2020;126:109903. https://doi.org/10.1016/j.biopha.2020.109903.
Ritchie ME, et al. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43: e47. https://doi.org/10.1093/nar/gkv007.
(2015) Gene Ontology Consortium: going forward. Nucleic acids research 43, D1049–1056, https://doi.org/10.1093/nar/gku1179.
Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. https://doi.org/10.1093/nar/28.1.27.
Yu G, Wang LG, Han Y, He QY. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. 2012;16:284–7. https://doi.org/10.1089/omi.2011.0118.
Subramanian A, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–50. https://doi.org/10.1073/pnas.0506580102.
Hänzelmann S, Castelo R, Guinney J. GSVA: gene set variation analysis for microarray and RNA-seq data. BMC Bioinform. 2013;14:7. https://doi.org/10.1186/1471-2105-14-7.
Langfelder P, Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinform. 2008;9:559. https://doi.org/10.1186/1471-2105-9-559.
Newman AM, et al. Determining cell type abundance and expression from bulk tissues with digital cytometry. Nat Biotechnol. 2019;37(7):773–82. https://doi.org/10.1038/s41587-019-0114-2.
Xiao B, et al. (2020) Identification and verification of immune-related gene prognostic signature based on ssGSEA for osteosarcoma. 10:607622.
Wakonig B, Auersperg AMI, O’Hara M. String-pulling in the Goffin’s cockatoo (Cacatua goffiniana). Learn Behav. 2021;49:124–36. https://doi.org/10.3758/s13420-020-00454-1.
Wang W, Zhang J, Wang Y, Xu Y, Zhang S. Identifies microtubule-binding protein CSPP1 as a novel cancer biomarker associated with ferroptosis and tumor microenvironment. Comput Struct Biotechnol J. 2022;20:3322–35. https://doi.org/10.1016/j.csbj.2022.06.046.
Chen Y, Wang X. miRDB: an online database for prediction of functional microRNA targets. Nucleic Acids Res. 2020;48:D127-d131. https://doi.org/10.1093/nar/gkz757.
Zhou KR, et al. ChIPBase v2.0: decoding transcriptional regulatory networks of non-coding RNAs and protein-coding genes from ChIP-seq data. Nucleic Acids Res. 2017;45:D43-d50. https://doi.org/10.1093/nar/gkw965.
Wagner M, Yoshihara M, Douagi I, Damdimopoulos A, Panula S, Petropoulos S, Lu H, Pettersson K, Palm K, Katayama S, Hovatta O, Kere J, Lanner F, Damdimopoulou P. Single-cell analysis of human ovarian cortex identifies distinct cell populations but no oogonial stem cells. Nat Commun. 2020;11(1):1147. https://doi.org/10.1038/s41467-020-14936-3.
Cohen-Fredarow A, Tadmor A, Raz T, Meterani N, Addadi Y, Nevo N, Solomonov I, Sagi I, Mor G, Neeman M, Dekel N. Ovarian dendritic cells act as a double-edged pro-ovulatory and anti-inflammatory sword. Mol Endocrinol. 2014;28(7):1039–54. https://doi.org/10.1210/me.2013-1400.
Wu R, Van der Hoek KH, Ryan NK, Norman RJ, Robker RL. Macrophage contributions to ovarian function. Hum Reprod Update. 2004;10(2):119–33. https://doi.org/10.1093/humupd/dmh011.
Turner EC, Hughes J, Wilson H, Clay M, Mylonas KJ, Kipari T, Duncan WC, Fraser HM. Conditional ablation of macrophages disrupts ovarian vasculature. Reproduction. 2011;141(6):821–31. https://doi.org/10.1530/REP-10-0327.
Chikuma S. CTLA-4, an Essential Immune-Checkpoint for T-Cell Activation. Curr Top Microbiol Immunol. 2017;410:99–126. https://doi.org/10.1007/82_2017_61.
Koyasu S. The role of PI3K in immune cells. Nat Immunol. 2003;4(4):313–9. https://doi.org/10.1038/ni0403-313.
Lawrence CP, Chow SC. Suppression of human T cell proliferation by the caspase inhibitors, z-VAD-FMK and z-IETD-FMK is independent of their caspase inhibition properties. Toxicol Appl Pharmacol. 2012;265(1):103–12. https://doi.org/10.1016/j.taap.2012.09.002.
Gaestel M, Kotlyarov A, Kracht M. Targeting innate immunity protein kinase signalling in inflammation. Nat Rev Drug Discov. 2009;8(6):480–99. https://doi.org/10.1038/nrd2829.
Barclay AN, Wright GJ, Brooke G, Brown MH. CD200 and membrane protein interactions in the control of myeloid cells. Trends Immunol. 2002;23(6):285–90. https://doi.org/10.1016/s1471-4906(02)02223-8.
Aref S, Azmy E, El-Gilany AH. Upregulation of CD200 is associated with regulatory T cell expansion and disease progression in multiple myeloma. Hematol Oncol. 2017;35(1):51–7. https://doi.org/10.1002/hon.2206.
Zhang D, Yu Y, Duan T, Zhou Q. The role of macrophages in reproductive-related diseases. Heliyon. 2022;8(11): e11686. https://doi.org/10.1016/j.heliyon.2022.e11686.
Al-Zeheimi N, Gao Y, Greer PA, Adham SA. Neuropilin-1 Knockout and Rescue Confirms Its Role to Promote Metastasis in MDA-MB-231 Breast Cancer Cells. Int J Mol Sci. 2023;24(9):7792. https://doi.org/10.3390/ijms24097792.
Szeliga A, Calik-Ksepka A, Maciejewska-Jeske M, Grymowicz M, Smolarczyk K, et al. Autoimmune Diseases in Patients with Premature Ovarian Insufficiency-Our Current State of Knowledge. Int J Mol Sci. 2021;22(5):2594. https://doi.org/10.3390/ijms22052594.
Funding
The work was supported by the Gansu Province Natural Science Foundation project, 22JR11RA010 and Gansu Province science and technology plan project, 23JRRA1321.
Author information
Authors and Affiliations
Contributions
Xiyan zhang wrote the manuscript and produced the figures and visualized the data. Luxiao Wei, Li Kong and Jing Du collected data,Ling Wang organized and revised the manuscript.Tongkun Yang revised the manuscript.All authors reviewed and approved the manuscript for publication.
Corresponding author
Ethics declarations
Conflict interest
The authors declare are no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Zhang, X., Wang, L., Yang, T. et al. Bioinformatic analysis of the role of immune checkpoint genes and immune infiltration in the pathogenesis and development of premature ovarian insufficiency. J Assist Reprod Genet (2024). https://doi.org/10.1007/s10815-024-03120-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s10815-024-03120-x