Abstract
Studies have shown that cultivars of the carrageenophytes Kappaphycus and Eucheuma are clones of a limited number of strains originally domesticated from wild populations. For the development and selection of new cultivars, it is important that a comprehensive record of available variants exists. This study was conducted to provide up-to-date analysis and compilation of the current state of cultivars as the last list of cultivars was compiled nearly a decade ago. The present study analyzed the cox2–3 spacer and cox1 (1356 bp) genetic diversity of cultivars collected from 2019 to 2020 from the east coast of Sabah where the seaweed farms are concentrated. These data were compared with cultivars reported from 2010 to 2012 to assess changes, if any, to the gene pool of farmed eucheumatoid in Malaysia. Kappaphycus alvarezii, K. striatus, and K. malesianus are currently cultivated while Eucheuma denticulatum is no longer an important cultivar compared to a decade ago, probably due to its lower price. Analysis of the cox2–3 spacer revealed a new haplotype, LBT10, and, by including published GenBank data, a further four previously unnamed haplotypes were recognized from Sabah. This study confirms that there is a limited gene pool within cultivars in Malaysia and suggests the need for new or genetically diverse cultivars which can adapt to a changing environment, to ensure a more sustainable carrageenan industry.
Similar content being viewed by others
Introduction
Kappaphycus and Eucheuma (herein referred to as eucheumatoids) are commercially important red (Rhodophyta) seaweeds valued for carrageenan, a useful phycocolloid especially in the food and cosmetic industries. The increasing demand for carrageenan has spurred on the commercial introduction of these rhodophytes to various parts of the world (Bixler and Porse 2011; FAO 2020; Brakel et al. 2021). In Malaysia, eucheumatoids are mainly cultivated on the east coast of Sabah which features long coastlines, extensive continental shelves, clean water, and being distant from estuaries and coral reefs (Sade et al. 2006; FAO 2018). Seaweed farming improves the social-economic conditions of the local coastal communities by offering job opportunities and income. As such, the development of the seaweed industry has always been one of the priority areas for the aquaculture industry, especially in Sabah (Hurtado et al. 2001; Sade et al. 2006; Phang et al. 2010; Nor et al. 2020). However, the cultivation practice in Malaysia has progressed little over the years, with production generally on a declining trend since a peak in 2012 (Online Resource 1). This is partly because the local farmers still rely on conventional farming techniques, including the use of the probably original K. alvarezii cultivar which has been vegetatively propagated for the past 50 years. The decline in production has probably also been affected by difficulties in generating healthy and vigorous seed stock, disease problems, and climate change (FAO 2018; Kumar et al. 2020; Ward et al. 2021). It is therefore crucial that Malaysia focus on the selection of good quality cultivars as a replacement for the declining strains. These qualities may include high carrageenan yield, fast growth, disease resistance, and heat tolerance among others. This can ultimately be achieved via marker-assisted selection (MAS), in which certain genetic markers correlate to traits of interest such as recent progresses seen in Saccharina japonica (Wang et al. 2018) and Pyropia yezoensis (Huang and Yan 2019; Yu et al. 2020).
Previous eucheumatoid genetic studies have shown the suitability of molecular markers in identifying species, inferring phylogeny, and assessing genetic variability (Zuccarello et al. 2006; Conklin et al. 2009; Tan et al. 2012, 2013; Barros-Barreto et al. 2013; Halling et al. 2013; Lim et al. 2014; Tano et al. 2015; Dumilag et al. 2016a, b, 2017; Thien et al. 2016, 2020; Roleda et al. 2021). The genetic characterization of Kappaphycus and Eucheuma in Southeast Asia using the cox1 gene and cox2–3 spacer by Lim et al. (2014) revealed numerous species and haplotypes of Kappaphycus that warranted further investigation for strain selection. However, the study raised concerns about the general lack of genetic diversity among the Kappaphycus and Eucheuma cultivars. Despite attempts to revisit the cultivation sites reported in Lim et al. (2014), seaweed farming has ceased at some of the sites (Sebangkat, Salakan, Sisipan, Pababag, Pangkor Island) for a variety of reasons, such as turtle and rabbitfish grazing and diseases (pers. comm. from farmers and fisheries officers). Nevertheless, since the collections made in 2010 to 2012, several new farming sites have been set up and collections were made from these areas including Umas-umas and Bust Point in Tawau (the latter close to the border with Indonesia) and new sites (Silungan, Silapang, Pallang-pallang, Ligit-ligitan, and Sangaban) near Semporna. Some farmers may change the location of farms seasonally or when production drops due to disease outbreaks (Brakel et al. 2021). As such, farming sites are likely to change over the years. Given the continuing decline in production since 2012, it is timely to reassess the genetic diversity of current cultivars used in Sabah as part of the measures to revitalize the industry. As this genetic information is deemed critical prior to any form of marker-assisted selection or cultivar development, the present study (i) assesses the cox1 and cox2–3 spacer genetic diversity of eucheumatoid cultivars from key farming localities in Sabah, Malaysia, and (ii) compares the genetic variation of these cultivars against those collected over the past decade.
Materials and methods
Eucheumatoid cultivars at various stages of culture (subject to availability at each farm) were collected from 13 farming sites in Semporna, Tawau, and Kunak, which are the top three production areas in Sabah (Fig. 1), from April 2019 to March 2020. Samples were selected by eye from the various colors and forms available at each farm at the time of collection with the intention to maximize the diversity of cultivars surveyed. For each species collected, the sample size ranged from 1 to 45 individuals, depending on their availability at each farm surveyed during the sampling period. Specimens were photographed and preserved as vouchers which were deposited in the University of Malaya Seaweeds and Seagrasses Herbarium (KLU). Depending on the sample size from each farm, one to three samples of each morphotype were selected accordingly for molecular analysis and the growing tips of each thallus were cut and dehydrated in silica gel. Details of the specimens examined in this study are summarized in Table 1.
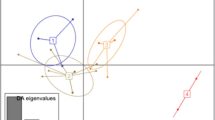
Map showing the sampling locations of cultivated eucheumatoids (depicted in shapes): 1, Madai; 2, Ampilan; 3, Ligit-ligitan; 4, Lok Buton; 5, Pallang-pallang; 6, Silapang; 7, Pelantar B Karindingan; 8, Pondohan Karindingan; 9, Sangaban; 10, Sipanggau; 11, Silungan; 12, Umas-umas; 13, Bust Point; 14, Pangkor Island; 15, Gaya Island; 16, Sandakan; 17, Kunak; 18, Sebangkat; 19, Omadal; 20, Salakan; 21, Sisipan; 22, Pababag. Circles denote collection sites of current study, triangles for sites from Lim et al. (2014), and diamond for site reported in Thien et al. (2020). Note that Thien et al. (2020) only mentioned Semporna (in general) and Pulau Gaya as the collection sites
Genomic DNA extraction and PCR amplifications followed Lim et al. (2014). Purified products were sequenced by Apical Scientific Sdn Bhd, Malaysia. The forward and reverse sequence reads were edited and assembled with ChromasPro v1.5 (Technelysium Pty. Ltd., Australia). Sequence data for both cox2–3 spacer and cox1 markers were generated for 74 cultivated eucheumatoids (Table 1). In addition, 88 cox2–3 spacer and 45 cox1 sequences of eucheumatoids were downloaded from GenBank (Online Resource 2), representing all recorded molecular diversity to date for Kappaphycus spp. and Eucheuma denticulatum. The sequences were aligned in ClustalX v. 2.0.8 (Larkin et al. 2007) and the alignments were manually revised and trimmed in Bioedit v. 7.0.9.0 (Hall 1999).
Haplotype networks were drawn in PopART v. 1.7 (http://popart.otago.ac.nz) (Leigh and Bryant 2015) by opting for the TCS network parameter with a default 95% connection limit and gaps as missing characters. DNA sequences with zero absolute and pairwise distance were collapsed into haplotypes and assigned the same haplotype code. Haplotype codes from previous studies were adopted for both phylogenetic and haplotype analyses (Zuccarello et al. 2006; Conklin et al. 2009; Tan et al. 2012; Barros-Barreto et al. 2013; Halling et al. 2013; Tan et al. 2013; Dumilag et al. 2016a, 2017). Haplotype analyses were performed separately for Kappaphycus spp. and E. denticulatum specimens. Haplotype network of the cox2–3 spacer was constructed and redrawn to visualize the relationships among haplotypes in each dataset. Only specimens with complementary cox2–3 spacer and cox1 (excluding cox1–5′) sequences were included in the concatenated dataset for analysis of mitochondrial haplotype diversity and frequency in Table 2.
Results
Overview of the dataset
During the sampling period from 2019 to 2020, only species of Kappaphycus (K. alvarezii, K. striatus, and K. malesianus) were cultivated in farms on the east coast of Sabah, Malaysia. Among these, K. alvarezii was the most widely cultivated species found in all three production areas (Semporna, Kunak, and Tawau). In contrast, the least cultivated species K. malesianus was only found in two cultivation sites in Semporna viz. Karindingan and Pallang-pallang. Sequences used in the phylogenetic trees were labeled according to their haplotypes (see Table 1 and Online Resource 2). Previously unrecognized haplotypes from Thien et al. (2020) were given codes and included in the analyses: K. alvarezii haplotypes SCK6 (cultivar from Semporna) and GCK3 (cultivar and wild from Semporna and Gaya Island in the northeast of Sabah), K. striatus haplotype SCK15 in cultivars and natural populations from Semporna and Gaya Island, and E. denticulatum haplotype SCE7 in cultivars and wild specimens from Semporna and Gaya Island.
Genetic diversity of eucheumatoid cultivars in Sabah, Malaysia
A total of 14 cox2–3 spacer haplotypes of K. alvarezii are now known to science (Fig. 2) and this includes the newly recognized haplotypes SCK6 and GCK3. One to three nucleotide differences separated the two new haplotypes from haplotype 3. In total, eight haplotypes are now recorded within K. striatus, of which five haplotypes have been found in Malaysia: in our dataset, cultivated K. striatus was dominated by haplotype 89 (29 out of 30 seaweeds), and we retrieved a new haplotype LBT10 (n = 1). Haplotype LBT10 differed from haplotype 89, which was also found in wild specimens from Malaysia and Indonesia, by one nucleotide each. The morphologies of LBT10 and 89 (Online Resource 3) are indistinguishable except for their color. All specimens of cultivated K. malesianus in this study were represented by a single haplotype for each dataset. None of the Malaysian cultivars that we collected in 2019–2020 belonged to Eucheuma. However, mining GenBank data enabled us to newly identify haplotype SCE7, which was found in cultivars and natural populations of E. denticulatum from Malaysia (Thien et al. 2020). This brings the total number of known cox2–3 spacer E. denticulatum haplotypes up to ten, of which three were previously observed in Malaysia (13, 32, and SCE7).
Statistical parsimony networks based on cox2–3 spacer for a Kappaphycus and b Eucheuma denticulatum. Each square box represents a haplotype. Haplotypes of the specimens collected in this study are indicated by asterisks and n = number of specimens. Each line between haplotypes indicates one mutation change. Missing haplotypes are indicated by small empty circles. The dashed circle denotes the new haplotype collected in this study and dashed boxes denote haplotypes newly recognized from published GenBank entries. Letters in square brackets indicate the collection status of the specimens, W = wild specimen and C = cultivated specimen. Letters in parentheses indicate countries of origin and abbreviated as follow: BR., Brazil; ID., Indonesia; MG., Madagascar; MU., Mauritius; MY., Malaysia; PH., Philippines; TZ., Tanzania; VN., Vietnam; SEA., Southeast Asia (Malaysia, Indonesia, Vietnam, and the Philippines); GL., global distribution
Comparison of genetic diversity within farmed eucheumatoids between 2010 and 2012 and 2019–2020
The number of haplotypes discovered within each species, haplotype codes, and abundance of each haplotype is summarized in Table 2. In 2019–2020, our sampling of K. alvarezii in Malaysian farms only retrieved cox2–3 spacer haplotype 3. In contrast, three haplotypes were found in cultivars collected from 2010 to 2012 with haplotype 3 as the most common cultivar (17 out of 26 seaweeds), followed by GCK3 (n = 7) and SCK6 (n = 2). With respect to concatenated haplotypes, KA was the most common cultivar reported in 2010–2012 (15 out of 16 seaweeds) while haplotype MY40 (n = 1) which had been found cultivated in Salakan, Sabah, by Lim et al. (2014) was not found again in this sampling campaign. Regarding K. malesianus cultivars, MY216 was the only haplotype observed (n = 7) since 2010 until 2020. The concatenated haplotypes reported earlier from wild populations in Malaysia viz. MY218 and MY92 have not apparently been introduced as cultivars, at least they were not found within our latest collections. The number of haplotypes for cultivated K. striatus in Malaysia declined from three (89, 117, and SCK15) in 2010–2012 to two (89, LBT10) in 2019–2020. Similar to the more recent results, haplotype 89 was the most common (6 out of 14 seaweeds) cultivar followed by SCK15 (n = 5) and 117 (n = 3). For E. denticulatum, haplotype 13 was the most common cultivar (n = 7) followed by SCE7 (n = 3) and 32 (n = 2) back in 2010 to 2012. Haplotype SCE7 was last reported in December 2010 from farm(s) in Semporna and a wild specimen from Gaya Island by Thien et al. (2020). Haplotypes 32, MY41, and EDB previously reported as cultivars have not been found since Lim et al. (2014).
Discussion
This study demonstrated a lack of mitochondrial haplotype diversity within Malaysian cultivars in the past 2 years, as only one haplotype each was found in K. alvarezii and K. malesianus, and two for K. striatus. Cultivars are also less diverse genetically compared to wild specimens. This is due to the repetitive vegetative propagation under a continuously active selection of the farmed stock (Ask and Azanza 2002). This scenario is not limited to Malaysia, but is also observed in neighboring Southeast Asian countries involved in eucheumatoid cultivation (Dumilag et al. 2016a; Ratnawati et al. 2020). A low level of genetic variation within cultivars potentially hinders their capacity for adaptation to environmental stress and disease resistance in the farming industry (Tano et al. 2015; Valero et al. 2017). While it is recognized that low mitochondrial diversity does not necessarily imply a limited overall genetic diversity and the inability to adapt, the absence of recombination especially from asexual propagation over the past five decades may have led to a gradual age-dependent fitness decay (Janko et al. 2008; Hollister et al. 2015). The use of the same cultivar strain affects the quality and overall productivity over time as they are more vulnerable to disease outbreak, particularly “ice-ice,” and epiphyte infestations which lead to significant losses to the stakeholders (Sade et al. 2006; Brakel et al. 2021).
The discovery of several novel haplotypes from Sabah based on published GenBank entries suggests the introduction of natural populations into culture, introduction of cultivars from elsewhere, or potential genetic pollution of cultivars into the wild. The origin of K. alvarezii cultivars with haplotypes SCK6 and GCK3 is unclear, particularly the latter which was found in both cultivars and wild specimens. The sampling extent and abundance of these haplotypes, along with SCK15 and SCE7, are unknown due to limited information in the original report by Thien et al. (2020). It is possible that the native population was introduced into farms for cultivation or that the cultivar introduced from elsewhere becomes an escapee from a farm. Despite the sharing of sea borders between Malaysia and its neighbors, specific cultivars reported only in Indonesia (e.g., INL5) and the Philippines (e.g., KALV-1 and KALV-2) have not been detected within farms in Malaysia. This may be a result of undersampling or that these haplotypes were rare and with locally restricted occurrence in the sites where they were first reported. Nevertheless, the movement of highly abundant strains across the sea borders is expected and this poses a biosecurity risk as diseases and pests may inadvertently be introduced from one place to another.
Like GCK3, the origin of SCK15 is unknown, although it was found in cultivars from Semporna and Gaya Island and a wild sample from Semporna. Haplotype 117 which was previously reported from natural populations (Lim et al. 2014) was later found in cultivars (Thien et al. 2020), indicating the possible introduction of wild genetic stock into the farms. There is also a possibility for wild samples of haplotype 117 being farm escapees that thrive in the natural habitats. Haplotypes MY220 and KSB previously reported in the wild have yet to be found within cultivars. In addition, cultivars used in Sitangkai, Tawi Tawi Philippines (SIT4 and SIT7), were not found in farms within Malaysian waters despite the proximity to Sabah. This may be due to their low abundance, one each out of 14 cultivated K. striatus reported in Lim et al. (2014). The absence of Eucheuma farming during our latest survey of eucheumatoid cultivation sites in Sabah for the past 2 years was unexpected and differed from a decade ago (Lim et al. 2014; Thien et al. 2020). In the past, both Kappaphycus and Eucheuma cultivars were encountered with the majority collected from the east coast of Sabah and a few K. alvarezii from Pangkor Island off the west coast of Peninsular Malaysia. Eucheuma produces iota-carrageenan which has a lower market demand and value over kappa-carrageenan produced by Kappaphycus species. This has inadvertently led to the cessation of E. denticulatum cultivation in Malaysia, as the seaweed farmers prefer to farm the higher-value Kappaphycus species with high carrageenan yield especially K. alvarezii.
A low accessible gene pool restricts the selection opportunity for strains well adapted to unfavorable environmental conditions (Halling et al. 2013). Therefore, it is important to increase the variation within the farmed stock by developing strains from the continuous selection of wild varieties and breeding programs to secure sustained production (Halling et al. 2013), in addition to preserving genetically distinct cultivars that are in use to maintain them as a source for breeding. Despite the morphological variations observed within the cultivars, morphological and genetic diversities may not have responded to the same environmental drivers (Cabrera-Toledo et al. 2020). Our results showed that the newly identified cultivars represented by haplotypes LBT10, SCK6, GCK3, and SCK15 could be promoted as potential alternatives to the dominant cultivars, subject to the assessment of their phenotypic qualities such as growth rate, carrageenan yield, and quality. The selection of strains from wild populations is also a potential means to diversify the number of cultivated genotypes, as they are a resource for new recombination with cultivated strains or between wild individuals for future genetic improvement of the crops. These highlight the urgent need for a regulated local biobank where (i) farmers can source their seedlings for cultivation instead of obtaining the seedlings indiscriminately from unregulated sources with implications for biosecurity and (ii) where the genetically distinct native germplasm, both currently available cultivars and natural populations, can be conserved for a sustainable seedstock supply.
In summary, the genetic diversity of eucheumatoid cultivars in Malaysia from 2010 to 2020 remains low despite the discovery of several new haplotypes. Concerningly, up to five previously reported haplotypes from cultivars were not found from recent sampling. This emphasizes the need to genotype currently cultivated strains and to ensure their conservation in preventing the loss of genetically distinct cultivars. Nevertheless, the genetic diversity of eucheumatoids in Malaysia is likely to be underestimated as there is insufficient sampling coverage for wild populations. Subsequent studies should place more focus on the wild eucheumatoids and the search for new markers, such as the whole genome or organellar genome sequencing, that can improve the resolution of intraspecific genetic variation within cultivars. Despite the lower demand for iota-carrageenan, farmers in Malaysia should consider the cultivation of E. denticulatum in tandem with Kappaphycus as the former has been reported to be less susceptible to ice-ice disease and epiphytes than Kappaphycus cultivars (Tisera and Naguit 2009; Pang et al. 2015). Research and development into improving or modifying the chemical properties of iota-carrageenan to suit the wide range of applications in the various industries may help to boost the demand for Eucheuma farming in the future.
Data Availability
All data generated or analyzed during this study are included in this published article and its supplementary information files.
References
Araújo PG, Miranda G, Barros-Barreto MB, Fujii M (2013) Molecular identification of the exotic lineage of Kappaphycus alvarezii (Rhodophyta, Solieriaceae) cultivated in the tropical region of Brazil. Phytotaxa 109:17–26
Ask EI, Azanza RV (2002) Advances in cultivation technology of commercial eucheumatoid species: a review with suggestions for future research. Aquaculture 206:257–277
Barros-Barreto MBB, Marinho LC, Reis RP, Mata CS, Ferreira PCG (2013) Kappaphycus alvarezii (Gigartinales, Rhodophyta) cultivated in Brazil: is it only one species? J Appl Phycol 25:1143–1149
Bast F, John AA, Bhushan S (2016) Molecular assessment of invasive carrageenophyte Kappaphycus alvarezii from India based on ITS-1 sequences. J Plant Taxon Geogr 71:287–292
Bindu MS, Levine IA (2010) The commercial red seaweed Kappaphycus alvarezii—an overview on farming and environment. J Appl Phycol 23:789–796
Bixler H, Porse H (2011) A decade of change in the seaweed hydrocolloids industry. J Appl Phycol 23:321–335
Brakel J, Sibonga RC, Dumilag RV, Montalescot V, Campbell I, Cottier-Cook EJ, Ward G, Le Masson V, Liu T, Msuya FE, Brodie J, Lim PE, Gachon CMM (2021) Exploring, harnessing and conserving marine genetic resources towards a sustainable seaweed aquaculture. Plants People Planet 3:337–349
Cabrera R, Umanzor S, Díaz-Larrea J, Araújo PG (2019) Kappaphycus alvarezii (Rhodophyta): new record of an exotic species for the Caribbean coast of Costa Rica. Am J Plant Sci 10:1888–1902
Cabrera-Toledo D, Vargas-Ponce O, Ascencio-Ramírez, Valadez-Sandoval LM, Pérez-Alquicira J, Morales-Saavedra J, Huerta-Galván OF (2020) Morphological and genetic variation in monocultures, forestry systems and wild populations of Agave maximiliana of Western Mexico: implications for its conservation. Front Plant Sci 11:817
Conklin KY, Kurihara A, Sherwood AR (2009) A molecular method for identification of the morphologically plastic invasive algal genera Eucheuma and Kappaphycus (Rhodophyta, Gigartinales) in Hawaii. J Appl Phycol 21:691–699
Dumilag RV (2018) Betaphycus gelatinus and B. philippinensis (Gigartinales, Rhodophyta) are conspecific. Phytotaxa 372:22–34
Dumilag RV, Orosco FL, Lluisma AO (2016a) Genetic diversity of Kappaphycus species (Gigartinales, Rhodophyta) in the Philippines. Syst Biodivers 14:441–451
Dumilag RV, Salvador RC, Halling C (2016b) Genotype introduction affects population composition of native Philippine Kappaphycus (Gigartinales, Rhodophyta). Conserv Genet Resour 8:439–441
Dumilag RV, Gallardo WGM, Garcia CPC, You YE, Chaves AKG, Agahan L (2017) Phenotypic and mtDNA variation in Philippine Kappaphycus cottonii (Gigartinales, Rhodophyta). Mitochondrial DNA A 29:951–963
FAO (2018) The global status of seaweed production, trade and utilization. Volume 124. Globefish Research Programme, Rome
FAO (2020) The state of the world fisheries and aquaculture 2020. Sustainability in action, Rome, p 244. https://doi.org/10.4060/ca9229en
Geraldino PJL, Yang EC, Boo SM (2006) Morphology and molecular phylogeny of Hypnea flexicaulis (Gigartinales, Rhodophyta) from Korea. Algae 21:417–423
Hall TA (1999) BioEdit: a user-friendly biological sequence alignment editor and analysis program for windows 95/98/NT. Nucleic Acids Symp Ser 41:95–98
Halling C, Wikström SA, Lilliesköld-Sjöö G, Mörk E, Lundsør E, Zuccarello GC (2013) Introduction of Asian strains and low genetic variation in farmed seaweeds: indications for new management practices. J Appl Phycol 25:89–95
Hollister JD, Greiner S, Wang W, Wang J, Zhang Y, Wong G-K, Wright SI, Johnson MTJ (2015) Recurrent loss of sex is associated with accumulation of deleterious mutations in Oenothera. Mol Biol Evol 32:896–905
Huang L, Yan X (2019) Construction of a genetic linkage map in Pyropia yezoensis (Bangiales, Rhodophyta) and QTL analysis of several economic traits of blades. PLoS One 14:e0209128
Hurtado AQ, Agbayani RF, Sanares R, Castro-Mallare MTR (2001) The seasonality and economic feasibility of cultivating Kappaphycus alvarezii in Panagatan cays, Caluya, antique, Philippines. Aquaculture 199:295–310
Janko K, Drozd P, Flegr J, Pannell JR (2008) Clonal turnover versus clonal decay: a null model for observed patterns of asexual longevity, diversity and distribution. Evolution 62:1264–1270
Jobb G, von Haeseler A, Strimmer K (2004) TREEFINDER: a powerful graphical analysis environment for molecular phylogenetics. BMC Evol Biol 4:18–26
Kumar YN, Poong S-W, Gachon C, Brodie J, Sade A, Lim P-E (2020) Impact of elevated temperature on the physiological and biochemical responses of Kappaphycus alvarezii (Rhodophyta). PLoS One 15:e0239097
Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG (2007) ClustalW and ClustalX version 2.0. Bioinformatics 23:2947–2948
Leigh J, Bryant D (2015) POPART: full-feature software for haplotype network construction. Methods Ecol Evol 6:1110–1116
Lim PE, Tan J, Phang SM, Nikmatullah A, Dang DH, Sunarpi H, Hurtado AQ (2014) Genetic diversity of Kappaphycus Doty and Eucheuma J. Agardh (Solieriaceae, Rhodophyta) in Southeast Asia. J Appl Phycol 26:1253–1272
Lim PE, Yang LE, Tan J, Maggs CA, Brodie J (2017) Advancing the taxonomy of economically important red seaweeds (Rhodophyta). Eur J Phycol 52:438–451
Nor AM, Gray TS, Caldwell GS, Stead SM (2020) A value chain analysis of Malaysia’s seaweed industry. J Appl Phycol 32:2161–2171
Pang T, Liu J, Liu Q, Li H, Li J (2015) Observations on pests and diseases affecting a eucheumatoid farm in China. J Appl Phycol 27:1975–1984
Paula EJ, Pereira R, Ohno M (1999) Strain selection in Kappaphycus alvarezii var. alvarezii (Solieriaceae, Rhodophyta) using tetraspore progeny. J Appl Phycol 11:111–121
Phang SM, Yeong HY, Lim PE, Nor ARM, Gan KT (2010) Commercial varieties of Kappaphycus and Eucheuma in Malaysia. Malays J Sci 29:214–223
Rambaut A, Drummond AJ (2009) FigTree version 1.3.1. Institute of Evolutionary Biology, University of Edinburgh, UK
Ratnawati P, Simatupang NF, Pong-Masak PR, Paul NA, Zuccarello GC (2020) Genetic diversity analysis of cultivated Kappaphycus in Indonesian seaweed farms using COI gene. Squalen Bull Mar Fish Postharv Biotech 15:65–72
Ronquist F, Huelsenbeck JP (2003) MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19:1572–1574
Sade A, Ali I, Mohd Ariff MR (2006) The seaweed industry in Sabah, East Malaysia. JATI 11:97–107
Swofford DL (2003) PAUP*. Phylogenetic analysis using parsimony (*and other methods). Version 4. Sinauer, Sunderland
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30:2725–2729
Tan J (2013) Molecular studies of Kappaphycus Doty and Eucheuma J. Agardh: Phylogenetics and DNA barcode assessment. PhD thesis, University of Malaya
Tan J, Lim PE, Phang SM, Hong DD, Sunarpi H, Hurtado AQ (2012) Assessment of four molecular markers as potential DNA barcodes for red algae Kappaphycus Doty and Eucheuma J. Agardh (Solieriaceae, Rhodophyta). PLoS ONE 7(12):e52905
Tan J, Lim PE, Phang SM (2013) Phylogenetic relationship of Kappaphycus Doty and Eucheuma J. Agardh (Solieraceae, Rhodophyta) in Malaysia. J Appl Phycol 25:13–29
Tan J, Lim PE, Phang SM, Rahiman AA, Nikmatullah A, Sunarpi H, Hurtado AQ (2014) Kappaphycus malesianus sp. nov.: a new species of Kappaphycus (Gigartinales, Rhodophyta) from Southeast Asia. J Appl Phycol 26:1273–1285
Tanabe AS (2007) Kakusan: a computer program to automate the selection of a nucleotide substitution model and the configuration of a mixed model on multilocus data. Mol Ecol Notes 7:962–964
Tano SA, Halling C, Lind E, Buriyo A, Wikström SA (2015) Extensive spread of farmed seaweeds causes a shift from native to non-native haplotypes in natural seaweed beds. Mar Biol 162:1983–1992
Thien VY, Yong WTL, Chin GJWL (2016) Morphological and molecular studies of undescribed Kappaphycus species. Int J Mar Sci 6:1–7
Thien VY, Yong WTL, Anton A, Chin GJWL (2020) A multiplex PCR method for rapid identification of commercially important seaweeds Kappaphycus alvarezii, Kappaphycus striatus and Eucheuma denticulatum (Rhodophyta, Solieriaceae). Reg Stud Mar Sci 40:101499
Tisera WL, Naguit MRA (2009) Ice-ice disease occurrence in seaweed farms in Bais Bay, Negros Oriental and Zamboanga del Norte. Threshold 4:1–16
Wang X, Chen Z, Li Q, Zhang J, Liu S, Duan D (2018) High-density SNP-based QTL mapping and candidate gene screening for yield-related blade length and width in Saccharina japonica (Laminariales, Phaeophyta). Sci Rep 8:13591
Ward G, Kambey C, Faisan Jr J, Tan P-L, Daumich C, Motoju I, Stentiford G, Bass D, Lim P-E, Brodie J, Poong S-W (2021) Ice-ice disease: an environmentally- and microbiologically driven syndrome in tropical seaweed aquaculture. Rev Aquacult. https://doi.org/10.1111/raq.12606
Yang EC, Kim MS, Geraldino PJL, Sahoo D, Shin J-A, Boo SM (2008) Mitochondrial cox1 and plastid rbcL genes of Gracilaria vermiculophylla (Gracilariaceae, Rhodophyta). J Appl Phycol 20:161–168
Yu X, Wang L, Xu K, Kong F, Wang D, Tang X, Sun B (2020) Fine mapping to identify the functional genetic locus for red coloration in Pyropia yezoensis thallus. Front Plant Sci 11:867
Zuccarello GC, Burger G, West JA, King RJ (1999) A mitochondrial marker for red algal intraspecific relationships. Mol Ecol 8:1443–1447
Zuccarello GC, Critchley AT, Smith J, Sieber V, Lhonneur GB, West JA (2006) Systematics and genetic variation in commercial Kappaphycus and Eucheuma (Solieriaceae, Rhodophyta). J Appl Phycol 18:643–651
Acknowledgements
The authors thank the officers of the Department of Fisheries Sabah (Mr. Maidin, Mr. Rosmin, Mr. Asim, Mr. Moksin, and Mr. Bidin), Mr. Japson Wong, Mr. Ghazali, Mr. Sazali, Miss Farah, and Miss Nurmaisyamimi, for their helpful assistance during sampling trips in Sabah, Malaysia.
Funding
This study is funded by UKRI through the GlobalSeaweedSTAR project (BB/P027806/1; IF015–2019), GlobalSeaweedSTAR Research Fund (RF002; IF059–2019), and by the Ministry of Higher Education Malaysia through the Higher Institution Centre of Excellence (HICoE) project (IOES-2014H).
Author information
Authors and Affiliations
Contributions
Conceptualization: Phaik-Eem Lim; methodology: Pui-Ling Tan; formal analysis and investigation: Pui-Ling Tan, Sze-Wan Poong; writing—original draft preparation: Pui-Ling Tan, Sze-Wan Poong, and Ji Tan; writing—review and editing: Janina Brakel, Claire Gachon, Juliet Brodie, Sze-Wan Poong, Ji Tan, Phaik-Eem Lim; funding acquisition: Phaik-Eem Lim, Claire Gachon, Juliet Brodie; resources: Ahemad Sade.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information

Online resource 1
Annual production (metric tons dry weight) of seaweeds in Malaysia from 2005 to 2020 (PNG 10374 kb)
Online resource 2
Details of Genbank eucheumatoid sequences used in this study (DOCX 34.1 kb)

Online resource 3
Morphological variants of cultivated K. alvarezii: a. PLF_1 “Tambalang” brown, b. MDI_11 “Tambalang” green, c. UMS_7 “Kangkung”, d. UMS_45 “Kangkung”, e. PDK_1 “Tambalang” brown, f. PDK_11 “Tambalang” green; K. striatus: g. LBT_2 “Flower” brown, h. LBT_10 “Flower”, i. LBT_17 “Flower”, j. LGT_1, k. LGT_6 “Red Durin”, l. SGB_3; K. malesianus: m. PLF_13 “Aring-aring”, n. PBK_30 “Aring-aring”, o. PDK_13 “Aring-aring”. Scale bars = 2 cm (PNG 67672 kb)
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Tan, PL., Poong, SW., Tan, J. et al. Assessment of genetic diversity within eucheumatoid cultivars in east Sabah, Malaysia. J Appl Phycol 34, 709–717 (2022). https://doi.org/10.1007/s10811-021-02608-8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10811-021-02608-8