Abstract
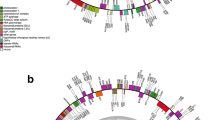
Betaphycus Doty, Eucheuma J. Agardh, and Kappaphycus Doty (Solieriaceae, Gigartinales) are the three most commercially important seaweed genera that produce carrageenan. In the present study we provide mitogenomes of Betaphycus gelatinus, Eucheuma denticulatum and Kappaphycus alvarezii. The mitogenomes of these three species contain a set of 50 genes, including 24 protein-coding genes, 2 rRNA genes, and 24 tRNA genes. The mitogenome length ranges from 25,198 bp (Kappaphycus alvarezii) to 25,327 bp (Eucheuma denticulatum). As compared with the previous published mitogenomes of Florideophyceae species, only the species in Gelidiaceae and Pterocladiaceae have smaller mitochondrial genome size than these reported here. At the junction of two transcription units, we identified a stem-loop structure in six representative Gigartinales species, which is presumed to play an important role in the replication and transcription of mitochondrial genes. In Gigartinales the difference in gene order among the four Solieriaceae (B. gelatinus, E. denticulatum, K. alvarezii, K. striatus) and other two Gigartinales species (Chondrus crispus and Mastocarpus papillatus) can be explained by inversion of two tRNA genes. Collinearity analysis of the 12 mitochondrial genomes of Florideophyceae showed considerable sequence synteny across all the species compared, with the exception of a highly variable region between atp6 and rpl20 genes. Phylogenetic analyses based on 21 shared mitochondrial genes showed that the four Solieriaceae species form one clade (Solieriaceae clade). Within this clade, B. gelatinae is basal relative to the other three species. The genus Kappaphycus is more closely related to Eucheuma than Betaphycus.




Similar content being viewed by others
References
Bixler HJ (1996) Recent developments in manufacturing and marketing carrageenan. Hydrobiologia 326:35–57
Bonen L (1998) Mitochondrial genomes. Adv Genome Biol 5:415–461
Boo GH, Hughey JR, Miller KA, Boo SM (2016) Mitogenomes from type specimens, a genotyping tool for morphologically simple species: ten genomes of agar-producing red algae. Sci Rep 6:35337
Boore JL, Brown WM (1998) Big trees from little genomes: mitochondrial gene order as a phylogenetic tool. Curr Opin Genet Dev 8:668–674
Burger G, Gray MW, Lang BF (2003) Mitochondrial genomes: anything goes. Trends Genet 19:709–716
Castresana J (2000) Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol 17:540–552
Clayton DA (1982) Replication of animal mitochondrial DNA. Cell 28:693–705
Conklin KY, Kurihara A, Sherwood AR (2009) A molecular method for identification of the morphologically plastic invasive algal genera Eucheuma and Kappaphycus (Rhodophyta, Gigartinales) in Hawaii. J Appl Phycol 21:691–699
Darling ACE, Mau B, Blattner FR, Perna NT (2004) Mauve: multiple alignment of conserved genomic sequence with rearrangements. Genome Res 14(7):1394–1403
Darriba D, Taboada GL, Doallo R, Posada D (2011) ProtTest 3: fast selection of best-fit models of protein evolution. Bioinformatics 27:1164–1165
Dierckxsens N, Mardulyn P, Smits G (2017) NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res 45:e18
Doty MS, Norris JN (1985) Eucheuma species (Solieriaceae, Rhodophyta) that are major sources of carrageenan. In: Abbott IA (ed) Taxonomy of economic seaweeds. California Sea Grant College Program, La Jolla, pp 47–61
Dumilag RV, Liao LM, Lluisma AO (2014) Phylogeny of Betaphycus (Gigartinales, Rhodophyta) as inferred from COI sequences and morphological observations on B. philippinensis. J Appl Phycol 26:587–595
Fredericq S, Freshwater DW, Hommersand MH (1999) Observations on the phylogenetic systematics and biogeography of the Solieriaceae (Gigartinales, Rhodophyta) inferred from rbcL sequences and morphological evidence. Hydrobiologia 398:25–38
Hall TA (1999) BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT; Nucl Acids Symp Ser 41:95–98
Heather JM, Chain B (2016) The sequence of sequencers: the history of sequencing DNA. Genomics 107:1–8
Huelsenbeck JP, Ronquist F (2001) MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 17:754–755
Leblanc C, Boyen C, Richard O, Bonnard G, Grienenberger JM, Kloareg B (1995) Complete sequence of the mitochondrial DNA of the rhodophyte Chondrus crispus (Gigartinales). Gene content and genome organization. J Mol Biol 250:484–495
Lim PE, Yang LE, Tan J, Meggs CA, Brodie J (2017) Advancing the taxonomy of economically important red seaweeds (Rhodophyta). Eur J Phycol 52:438–451
Lohse M, Drechsel O, Bock R (2007) OrganellarGenomeDRAW (OGDRAW): a tool for the easy generation of high-quality custom graphical maps of plastid and mitochondrial genomes. Curr Genet 52:267–274
Ma PF, Zhang YX, Zeng CX, Guo ZH, Li DZ (2014) Chloroplast phylogenomic analyses resolve deep-level relationships of an intractable bamboo tribe Arundinarieae (Poaceae). Syst Biol 63:933–950
Martin WF, Müller M (1998) The hydrogen hypothesis for the first eukaryote. Nature 392:37–41
Ng PK, Lin SM, Lim PE, Liu LC, Chen CM, Pai TW (2017) Complete chloroplast genome of Gracilaria firma (Gracilariaceae, Rhodophyta), with discussion on the use of chloroplast phylogenomics in the subclass Rhodymeniophycidae. BMC Genomics 18:40
Pereira L, Meireles F, Abreu HT, Ribeiro-Claro PJA (2015) A comparative analysis of carrageenans produced by underutilized versus industrially utilized macroalgae (Gigartinales, Rhodophyta). In: Kim S-K, Chojnacka K (eds) Marine algae extracts: processes, products, and applications. Wiley-VCH, Weinheim, pp 277–294
Schattner P, Brooks AN, Lowe TM (2005) The tRNAscan-SE, snoscan and snoGPS web servers for the detection of tRNAs and snoRNAs. Nucleic Acids Res 33:686–689
Sissini MN, Navarrete-Fernández TM, Murray EMC, Freese JM, Gentilhomme AS, Huber SR, Mumford TF, Hughey JR (2016) Mitochondrial and plastid genome analysis of the heteromorphic red alga Mastocarpus papillatus (C. Agardh) Kützing (Phyllophoraceae, Rhodophyta) reveals two characteristic florideophyte organellar genomes. Mitochondrial DNA B 1:676–677
Stamatakis A (2006) RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22:2688–2690
Sun YY, Luo D, Zhao C, Li W, Liu T (2011) DNA extraction and PCR analysis of five kinds of large seaweed under different preservation conditions. Mol Plant Breed 9:1680–1691
Taanman JW (1999) The mitochondrial genome: structure, transcription, translation and replication. Biochim Biophys Acta 1410:103–123
Tablizo FA, Lluisma AO (2014) The mitochondrial genome of the red alga Kappaphycus striatus (“Green Sacol” variety): complete nucleotide sequence, genome structure and organization, and comparative analysis. Mar Genomics 18:155–161
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28:2731–2739
Tan J, Lim P-E, Phang S-M (2013) Phylogenetic relationship of Kappaphycus Doty and Eucheuma J. Agardh (Solieriaceae, Rhodophyta) in Malaysia. J Appl Phycol 25:13–29
Tirtawijaya G, Mohibbullah M, Meinita MDN, Moon IS, Hong Y-K (2016) The ethanol extract of the rhodophyte Kappaphycus alvarezii promotes neurite outgrowth in hippocampal neurons. J Appl Phycol 28:2515–2522
Vellai T, Takacs K, Vida G (1998) A new aspect to the origin and evolution of eukaryotes. J Mol Evol 46:499–507
Yang Z (2007) PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol 24:1586–1591
Yang EC, Kim KM, Kim SY, Lee J, Boo GH, Lee J-H, Nelson WA, Yi G, Schmidt WE, Fredericq S, Boo SM, Bhattacharya D, Yoon HS (2015) Highly conserved mitochondrial genomes among multicellular red algae of the Florideophyceae. Genome Biol Evol 7:2394–2406
Zhang L, Wang X, Qian H, Chi S, Liu C, Lui T (2012) Complete sequences of the mitochondrial DNA of the wild Gracilariopsis lemaneiformis and two mutagenic cultivated breeds (Gracilariaceae, Rhodophyta). PLoS One 7(6):e40241
Zuccarello GC, Critchley AT, Smith J, Sieber V, Lhonneur GB, West JA (2007) Systematics and genetic variation in commercial Kappaphycus and Eucheuma (Solieriaceae, Rhodophyta). In: Anderson R, Brodie J, Onsøyen E, Critchley AT (eds) Proceedings of the Eighteenth International Seaweed Symposium. Springer, Dordrecht, pp 417–425
Funding
This work was supported by the China-ASEAN Maritime Cooperation Fund, by Public Science and Technology Research Funds Projects of Ocean (Grant No. 201405020), and by the Fundamental Research Funds for the Central Universities.
Author information
Authors and Affiliations
Corresponding authors
Rights and permissions
About this article
Cite this article
Li, Y., Liu, N., Wang, X. et al. Comparative genomics and systematics of Betaphycus, Eucheuma, and Kappaphycus (Solieriaceae: Rhodophyta) based on mitochondrial genome. J Appl Phycol 30, 3435–3443 (2018). https://doi.org/10.1007/s10811-018-1450-1
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10811-018-1450-1