Abstract
Purpose
To identify the spectrum of RB1 gene mutations in 114 Chinese patients with retinoblastoma.
Methods
Genomic DNA was extracted from the peripheral blood of 114 Rb patients. Polymerase chain reactions (PCRs) followed by direct Sanger sequencing were used to screen for mutations in the RB1 gene, which contains 26 exons with flanking intronic sequences, except exon 15. Clinical data, including gender, age at diagnosis, laterality of ocular lesions, and associated symptoms, were recorded and compared.
Results
We identified five novel mutations in the RB1 gene. Twenty-five other mutations found in this study have been previously reported. A higher rate of RB1 mutations, with 47.3% of mutations among bilaterally affected patients vs. 6.8% within unilaterally affected patients, was also observed (p < 0.0001). Bilaterally affected patients were diagnosed earlier when compared to unilaterally affected patients (11 ± 7 months versus 20 ± 14 months, p = 0.0002). Furthermore, nonsense mutations were abundant (n = 14), followed by frameshift mutations (n = 8), splicing site mutations (n = 5), while missense mutations were few (n = 3).
Conclusions
We found five novel mutations in RB1 genes, which expands the mutational spectrum of the gene. Children with bilateral Rb exhibited higher mutation rates and were diagnosed earlier than those with unilateral Rb. These findings will inform clinical diagnosis and genetic therapeutic targeting in Rb patients.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Retinoblastoma (Rb; OMIM 180200), the most common primary intraocular malignancy in children, has a worldwide incidence rate of 1:15,000—20,000 live births [1]. About 95% of Rb cases are diagnosed before the age of five [2]. In developed countries, due to effective modern treatment, this disease is associated with a lower mortality rate and improved outcomes. In contrast, in developing countries, due to lack of prompt diagnosis and effective treatment, Rb patients have a 40–70% mortality rate and an increased risk of enucleation [3]. In addition, Rb survivors have higher risks of secondary tumors [4], including bone and soft tissue sarcomas, melanomas, leukemia, lung cancer, uterine leiomyosarcoma, and radiotherapy-related central nervous system tumors [5]. Secondary tumors adversely affect the quality of life and survival rate of Rb patients.
Mutations in RB1, the first described tumor suppressor gene located on chromosome 13q14.2, have been associated with heritable Rb [6, 7]. Mutated RB1 gene may result in a loss of function of the retinoblastoma protein (pRB). Moreover, pRB knockdown was shown to promote tumor cell proliferation and repressed E2F activities [8]. Based on Knudson’s two-hit hypothesis, Rb development is mainly caused by mutations of both RB1 alleles [9]. Individuals with RB1 germline mutations are genetically predisposed to Rb [3]. Moreover, the types and frequencies of mutated RB1 in children developing Rb vary geographically. Therefore, identification of RB1 (Gene ID:5925; NM_000321) is key in genetic Rb testing and screening [10]. Genetic testing of the RB1 gene is inevitable for early diagnosis in heritable Rb to provide timely therapies and improve prognosis [11]. However, in China, limited data are available on RB1 mutations for prompt screening of all RB1 mutations, avoiding false negatives in genetic testing of Rb patients, and informing the provision of effective therapies.
In this study, we characterized the spectrum of RB1 mutations in children with Rb in China. Furthermore, the data generated in this study will form the basis for effective genetic counseling of Rb patients and their families, thereby improving Rb diagnosis and prognosis.
Material and methods
Patients
This study included 114 patients with retinoblastoma from Southern China. Clinical diagnosis of retinoblastoma was based on the clinical features and confirmed by an expert pathologist. All patients with Rb were diagnosed and treated according to the latest China guidelines for the diagnosis and treatment of retinoblastoma. Clinical data of all patients were collected from the hospital information system, including age, gender, tumor laterality, age at diagnosis, and associated symptoms (Table 1). All participants gave their written informed consent. The study was approved by the Ethics Committee of The Fifth Affiliated Hospital, Sun Yat-sen University, and was conducted in accordance with the principles of the 1975 Declaration of Helsinki.
Mutation detection
PCR
Genomic DNA was extracted from blood samples of patients firstly diagnosed with Rb using the iPure blood genomic DNA kit (IGEbio, CZ313-S). The extracted DNA was quantified using Nanodrop 2000 (Nanodrop, Wilmington, DE, USA). The primers used for detecting RB1 mutations (shown in Supplementary Table S1) were designed using Primer Premier 3.0 software for 26 translating exons and the adjacent intron–exon regions of the RB1 gene, except exon 15. The PCR had a total volume of 20 µl, including 10 µl HiFipfu polymerase (IGEbio, P300-S), 7 µl nuclease-free water, 1 µl forward and reverse primers (10 uM) each, and 2 µl DNA template. A BIO-RAD T100 TM Thermal cycler (Bio-Rad, Gladesville, Australia) and a touchdown PCR protocol were used for amplification. The PCR conditions were as follows: initial denaturation at 94 °C for 5 min followed by 10 cycles of denaturation at 94 °C for 30 s, annealing at 65 °C for 30 s, with the temperature of the heating block decreased by 1 °C per cycle, and then extension at 72 °C for 40 s. This was then followed by 25 cycles of denaturation at 94 °C for 30 s, annealing at 55 °C for 30 s, extension at 72 °C for 40 s and final extension at 72 °C for 3 min.
Sanger sequencing and sequence analysis
The PCR products were visualized by 1.5% agarose gel electrophoresis. The target band was excised, after which the DNA was purified and sequenced with ABI3730XL (Applied Biosystems, Foster City, CA, USA) using the BigDye method. Nucleotide sequences of the RB1 gene (GenBank L11910.1) were used as the reference. A Sequence Scanner, version 1.0 (Applied Biosystems, Streetsville, Ontario, Canada), and chromas (2.6.5) were used to align each exon’s sequence with the reference sequence to annotate mutations. Variants were described based on the latest nomenclature for the description of sequence variants (Human Genome Variation database: http://www.HGVS.org/varnomen). Additional information on RB1 gene mutations was obtained from Clinvar (https://www.ncbi.nlm.nih.gov/clinvar/), Human Gene Mutation Database (HGMD: http://www.hgmd.cf.ac.uk/ac/), LOVD (http://www.lovd.nl/3.0/home), gnomAD (http://gnomad.broadinstitute.org/), ensemble, and dbSNP. pRB protein structures were retrieved from UniProt (http://www.uniprot.org/uniprot, P06400). Open reading frame (ORF) and frameshift mutation predictions were performed using the NCBI ORF finder (https://www.ncbi.nlm.nih.gov/orffinder). The pathogenicity of variants was interpreted according to ACMG guidelines.
Statistical analysis
Student’s t test was performed to compare differences between continuous variables. Categorical variables were compared using Chi-square or Fisher’s exact tests. p ≤ 0.05 was considered statistically significant. Data analysis was performed using the SPSS software, version 25.0.
Results
Clinical characteristics
A total of 114 Chinese patients with Rb were recruited for this study, including 68 males and 46 females (59.6% vs 40.4%). A summary of the cases and their clinical characteristics are shown in Table 1. Fifty-five patients were bilaterally affected, while fifty-nine patients were unilaterally affected (48.2% versus 51.8%). In detail, unilateral cases were observed in 40 males and 19 females, while bilateral cases were observed in 28 males and 27 females (p = 0.0859). In unilateral cases, distributions of left and right ocular lesions were relatively similar (30 versus 29, respectively). The age at diagnosis ranged from 1 to 64 months old, with an average age of 14 months. Moreover, 91 patients (79.8%) had Rb onset before the age of 2 years, while 23 patients (20.2%) had Rb onset after 2 years of age. Most of the bilaterally affected patients (53/55, 96.4%) were diagnosed before 2 years. Mean age at diagnosis for patients with bilateral Rb was lower than for unilateral cases (11 ± 7 months versus 20 ± 14, p < 0.0001). This finding is consistent with those of previous studies [3, 12, 13]. Leukocoria was the most common initial symptom in Rb cases, accounting for 65.8% of all cases, followed by squint (21.9%). Several patients presented with other symptoms, including redness, accompanied by tearing, vision diminution, calcification, and white spots.
Mutations
Novel mutations
A total of five novel mutations were identified in our Rb cases. Sequencing analysis of the five novel variants is presented in Fig. 1. There were four frameshift mutations and one splice site mutation consisting of c.180_187del, c.528del, c.2035_2039del, c.2299_2300del, and c.1050-2A > T. All mutations were identified in bilaterally affected Rb patients with a mean age at diagnosis of 13.6 months. The splice site mutation, c.1050-2A > T, presenting at 3’ splice site of intron 10, altered the sequence from AG to TG and disrupted the canonical splice site. Intriguingly, the four novel frameshift mutations were predicted to result in a truncated RB protein arising from a premature stop codon in the reading frame and the loss of coding exons. Furthermore, three of the four frameshift mutations resulted in premature termination of amino acid translation in the conserved pRB pocket region, encoding transcriptional regulation and forming the repressor motif [14]. As shown in Fig. 2, c.180_187del (p.Cys61Ilefs*46) and c.528del (p.Gln176Hisfs*10) mutations occurred in exon2 and exon5, causing a frameshift variant and a premature stop codon at amino acid 106 and 185, and truncating 822 and 743 amino acids, respectively, at the C terminus of the RB1 protein. In addition, c.2035_2039del (p.Ile679Leufs*11) was located at domain B of the pRB pocket, resulting in premature translational termination at position 689, subsequently truncating 239 amino acids of the predicted protein. The other novel frameshift mutation, c.2299_2300del (p.Asn767Tyrfs*27), generated a frameshift and premature termination of the open reading frame.
Sequencing chromatograms illustrating novel mutations in the patients. The left panels show the mutations identified in Rb patients, while the right panels show the wildtype RB1 sequences. Arrow indicates the mutated site. A c.180_187del heterozygous mutation in patient Rb12. B c.528del heterozygous mutation in patient Rb119. C c.2035_2039del heterozygous mutation in patient Rb47. D c.1050-2A > T heterozygous mutation in patient Rb7. E c.2299_2300del heterozygous mutation in patient Rb81
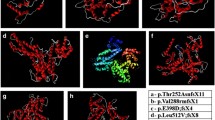
Effects of the four novel frameshift mutations on the RB1 protein. c.180_187del (p.Cys61Ilefs*46) mutations caused a frameshift change and a premature stop codon at amino acid 106, truncating 822 amino acids at the C terminus of the RB1 protein. c.528del (p.Gln176Hisfs*10) mutations caused a frameshift change and a premature stop codon at amino acid 185, truncating 743 amino acids at the C terminus of the RB1 protein. c.2035_2039del (p.Ile679Leufs*11) mutations caused a frameshift change and a premature stop codon at amino acid 689, truncating 239 amino acids at the C terminus of the RB1 protein. c.2299_2300del (p.Asn767Tyrfs*27) mutations caused a frameshift change and a premature stop codon at amino acid 793, truncating 135 amino acids at the C terminus of the RB1 protein
Other mutations
Twenty-five other previously reported RB1 mutations we found in this study are shown in Table 2. The most common mutational type was nonsense mutations (fourteen of twenty-five; 56.0%), followed by frameshift (three of twenty-five; 12.0%), splice mutations (three of twenty-five; 12.0%), and missense mutation (three of twenty-five, 12.0%).
As shown in Table 3, in a total of 30 variants, the most commonly observed RB1 gene alteration was nonsense mutation, accounting for 46.7% of the detected mutations, followed by frameshift mutations (26.6%), splicing alterations (16.7%), and missense mutations (10.0%). Mutations located at exons 8, 18, 20 exhibited the highest mutation frequencies and were, respectively, found in 3 patients. Mutations at exons 1, 13, 14, 17, 22, and intron 21 were, respectively, found in 2 patients. There were no mutations in exons 6, 7, 9, 11, 16, 22, and 24–27. Furthermore, 50.0% (15/30) of the detected mutations were located at regions encoding two conserved domains, A (residue 373–573) and B (residue 646–765), of the pRB pocket containing five nonsense, four splicing, four frameshift, and two missense variants. Specifically, 4/5 splice site mutations and 2/3 missense mutations were located at the pRB pocket region. The mutation rate in domain A located at exons 12–18 was 26.7% (8/30), while the mutation rate in domain B located at exons 19–23 was 23.3% (7/30). In addition, 21/30 mutations caused a premature stop codon on RNA transcripts resulting in a truncated protein.
Of the 14 cases with nonsense mutations, nine (64.3%) patients had C to T transitions in CGA codons in exons 8, 10, 14, 18, with several of these mutations affecting more than two unrelated patients. For instance, the c.1735C > T (p.Arg579*) mutation in exon 18 was detected in three unrelated patients, while the c.763C > T (p.Arg255*) mutation in exon8 was detected in two unrelated patients. Additionally, c.2242G > T(p.Glu748*) was detected in the proband’s mother, who did not suffer from Rb, indicating that this mutation was maternally inherited. Four of the eight reported frameshift mutations were novel mutations that had never been previously reported or characterized in mutated RB1. Moreover, four of the five splicing mutations occurred by substitution of G to A/T. Cases with splice site mutation were diagnosed at a younger age, ranging from 1 to 8 months.
Genotype–phenotype correlations
Sequencing analysis was performed for the 26 exons and adjacent intronic regions, except exon 15. DNA analysis revealed 27 different causative RB1 mutations in 30 patients (26.3%), which were identified in 4/59 (6.8%) patients with unilateral Rb and 26/55 (47.3%) patients with bilateral Rb (p < 0.0001) (Fig. 3A). The mutation rate in the bilaterally affected patients was significantly higher than in the unilaterally affected patients, consistent with previous findings [15]. Mean age at diagnosis for patients with mutated RB1 was 13 ± 11 months and 16 ± 12 months for patients without mutated RB1. Differences in age at diagnosis between the two groups were not significant (p = 0.1807) (Fig. 3B).
Mutation detection rate and mean age at diagnosis in retinoblastoma patients. A The left pie chart shows an overall mutation detection rate of 26.3% (30/114) in all Rb cases. The middle pie chart shows the mutation rate of 47.3% (26/30) in bilateral Rb cases. The right panel shows a mutation detection rate of 6.8% (4/59) in unilateral Rb cases. B Age at diagnosis of probands with positive (n = 30) or negative (n = 84) RB1 mutation test results
Discussion
In this study, we identified RB1 gene mutations in 26.3% of the 114 recruited Chinese Rb patients. This included five novel mutations that had never been previously reported. Bilaterally affected Rb patients exhibited higher mutation rates and an earlier age at diagnosis than unilaterally affected patients.
Most frameshift or nonsense mutations in inherited diseases or cancers result in premature termination codons (PTCs) [16]. In this study, nonsense and frameshift mutations were the major mechanisms of RB1 inactivation, accounting for 73.3% of all RB1 mutations. This finding is consistent with previously reported rates of 50–80.32% [4, 12, 17,18,19]. Furthermore, 50.0% of all RB1 mutations lie in the A/B “pocket” domain of pRB, which can bind E2F transcription factors and promote protein–protein interactions [20]. This mutation rate was comparable to previously reported rates of 40–78.95% [4, 12, 17, 21, 22]. In addition, subtle mutations may result in partial inactivation of pRB, decreasing protein stability. Therefore, variants occurring in or adjacent to known conserved protein domains have a great influence on disease onset [23].
Premature stop codons resulting in loss-of-function (LOF) alleles are strongly associated with inactivation of the RB protein [24]. Accordingly, frameshift mutations with premature translation termination might contribute to the onset of diseases by degrading RB1 transcripts, resulting in the complete loss of pRb. In this study, four of the five novel RB1 variants were frameshift mutations and were reported in patients with bilateral lesions. According to the ACMG standards and guidelines [25], all novel RB1 frameshift variants were pathogenic. This finding supports the postulate that frameshift mutations are highly associated with retinoblastoma disease. Studies should evaluate the specific impacts of these novel mutations on protein functions.
A marginally lower degree of RB1 germline mutation rate was reported in this study, compared to studies done in other countries [17,18,19, 26, 27]. Our findings are consistent with several studies conducted in China, with 21–27.1% of total RB1 mutation rates detected using Sanger sequencing alone [7, 28, 29]. Therefore, we hypothesized that geographical variations or race accounted for these differences. The heterogeneity in mutation detection rates can also be explained by the different techniques, such as MLPA and next-generation sequencing [30], used to detect mutations. In addition, the fact that we missed sequencing analysis of the promoter region and exon 15 due to the poly (A) and poly (T) sequence on both sides could also explain this difference.
Retinoblastoma is a highly complex disease. Single-nucleotide variants, large rearrangements, and promoter hypermethylation of the RB1 gene have a role in its development [31, 32]. Furthermore, low-level mosaic and deep intronic variants have been identified [33]. Besides, other genes may also be attributed to the Rb phenotype. For instance, MED4, a synthetic lethal target in tumors, explains the low penetrance of retinoblastoma [34]. In addition, amplification of the MYCN oncogene contributes to retinoblastoma [35]. Besides, retinoblastoma has also been associated with MDM2, the first modifier gene so far identified in retinoblastoma [36]. Further, the PIN1 gene that alters pRB phosphorylation is also important for Rb development [37]. Despite extensive research, the exact pathomechanisms of Rb remain elusive and should be investigated further. Given the relatively high negative rate of RB1 mutations in this study, we will perform whole-exome sequencing (WES) in samples obtained from patients without RB1 mutations to explore other possible disease-causing genes and variants. We postulate that our findings will elucidate the pathogenesis of Rb.
The spectrum of RB1 mutations in this study will facilitate the development of better-targeted therapies and inform on optimal disease management as well as effective treatment of Rb patients [38]. Prenatal diagnosis and family screening should be performed to identify individuals with a genetic predisposition to Rb [39].
Data availability
The findings of this study are included within the article and are available from the corresponding author upon reasonable request.
Abbreviations
- Rb:
-
Retinoblastoma
- PTCs:
-
Premature termination codons
- LOF:
-
Loss of function
- MLPA:
-
Multiplex ligation-dependent probe amplification
- WES:
-
Whole-exome sequencing
References
Fabian ID, Onadim Z, Karaa E, Duncan C, Chowdhury T, Scheimberg I, Ohnuma SI, Reddy MA, Sagoo MS (2018) The management of retinoblastoma. Oncogene 37(12):1551–1560. https://doi.org/10.1038/s41388-017-0050-x
Avior Y, Lezmi E, Yanuka D, Benvenisty N (2017) Modeling developmental and tumorigenic aspects of trilateral retinoblastoma via human embryonic stem cells. Stem Cell Reports 8(5):1354–1365. https://doi.org/10.1016/j.stemcr.2017.03.005
Dimaras H, Kimani K, Dimba EA, Gronsdahl P, White A, Chan HS, Gallie BL (2012) Retinoblastoma. Lancet 379(9824):1436–1446. https://doi.org/10.1016/s0140-6736(11)61137-9
Tomar S, Sethi R, Sundar G, Quah TC, Quah BL, Lai PS (2017) Mutation spectrum of RB1 mutations in retinoblastoma cases from Singapore with implications for genetic management and counselling. PLoS ONE 12(6):e0178776. https://doi.org/10.1371/journal.pone.0178776
Chai P, Luo Y, Yu J, Li Y, Yang J, Zhuang A, Fan J, Han M, Jia R (2021) Clinical characteristics and germline mutation spectrum of RB1 in Chinese patients with retinoblastoma: A dual-center study of 145 patients. Exp Eye Res 205:108456. https://doi.org/10.1016/j.exer.2021.108456
Chen CY, Xu CM, Du ZF, Chen XL, Ren GL, Zhang XN (2010) A c.1363C>T (p.R455X) nonsense mutation of RB1 gene in a southern Chinese retinoblastoma pedigree. Genet Test Mol Biomarkers 14(2):193–196. https://doi.org/10.1089/gtmb.2009.0162
He MY, An Y, Gao YJ, Qian XW, Li G, Qian J (2014) Screening of RB1 gene mutations in Chinese patients with retinoblastoma and preliminary exploration of genotype-phenotype correlations. Mol Vis 20:545–552
Sahin F, Sladek TL (2010) E2F–1 has dual roles depending on the cell cycle. Int J Biol Sci 6(2):116–128. https://doi.org/10.7150/ijbs.6.116
Knudson AG Jr (1971) Mutation and cancer: statistical study of retinoblastoma. Proc Natl Acad Sci U S A 68(4):820–823. https://doi.org/10.1073/pnas.68.4.820
Soliman SE, Racher H, Zhang C, MacDonald H, Gallie BL (2017) Genetics and Molecular Diagnostics in Retinoblastoma–An Update. Asia Pac J Ophthalmol (Phila) 6(2):197–207. https://doi.org/10.22608/apo.201711
Zou Y, Li J, Hua P, Liang T, Ji X, Zhao P (2021) Spectrum of germline mutations in RB1 in Chinese patients with retinoblastoma: application of targeted next-generation sequencing. Mol Vis 27:1–16
Lan X, Xu W, Tang X, Ye H, Song X, Lin L, Ren X, Yu G, Zhang H, Wu S (2020) Spectrum of RB1 germline mutations and clinical features in unrelated Chinese patients with retinoblastoma. Front Genet 11:142. https://doi.org/10.3389/fgene.2020.00142
MacCarthy A, Birch JM, Draper GJ, Hungerford JL, Kingston JE, Kroll ME, Onadim Z, Stiller CA, Vincent TJ, Murphy MF (2009) Retinoblastoma in Great Britain 1963–2002. Br J Ophthalmol 93(1):33–37. https://doi.org/10.1136/bjo.2008.139618
Chow KN, Dean DC (1996) Domains A and B in the Rb pocket interact to form a transcriptional repressor motif. Mol Cell Biol 16(9):4862–4868. https://doi.org/10.1128/mcb.16.9.4862
Nyamori JM, Kimani K, Njuguna MW, Dimaras H (2012) The incidence and distribution of retinoblastoma in Kenya. Br J Ophthalmol 96(1):141–143. https://doi.org/10.1136/bjophthalmol-2011-300739
Frischmeyer PA, Dietz HC (1999) Nonsense-mediated mRNA decay in health and disease. Hum Mol Genet 8(10):1893–1900. https://doi.org/10.1093/hmg/8.10.1893
Nguyen HH, Nguyen HTT, Vu NP, Le QT, Pham CM, Huyen TT, Manh H, Pham HLB, Nguyen TD, Le HTT, Van Nong H (2018) Mutational screening of germline RB1 gene in Vietnamese patients with retinoblastoma reveals three novel mutations. Mol Vis 24:231–238
Rojanaporn D, Boontawon T, Chareonsirisuthigul T, Thanapanpanich O, Attaseth T, Saengwimol D, Anurathapan U, Sujirakul T, Kaewkhaw R, Hongeng S (2018) Spectrum of germline RB1 mutations and clinical manifestations in retinoblastoma patients from Thailand. Mol Vis 24:778–788
Kiet NC, Khuong LT, Minh DD, Quan NHM, Xinh PT, Trang NNC, Luan NT, Khai NM, Vu HA (2019) Spectrum of mutations in the RB1 gene in Vietnamese patients with retinoblastoma. Mol Vis 25:215–221
Balog ER, Burke JR, Hura GL, Rubin SM (2011) Crystal structure of the unliganded retinoblastoma protein pocket domain. Proteins 79(6):2010–2014. https://doi.org/10.1002/prot.23007
Price EA, Price K, Kolkiewicz K, Hack S, Reddy MA, Hungerford JL, Kingston JE, Onadim Z (2014) Spectrum of RB1 mutations identified in 403 retinoblastoma patients. J Med Genet 51(3):208–214. https://doi.org/10.1136/jmedgenet-2013-101821
Sagi M, Frenkel A, Eilat A, Weinberg N, Frenkel S, Pe’er J, Abeliovich D, Lerer I (2015) Genetic screening in patients with Retinoblastoma in Israel. Fam Cancer 14(3):471–480. https://doi.org/10.1007/s10689-015-9794-z
Dyson NJ (2016) RB1: a prototype tumor suppressor and an enigma. Genes Dev 30(13):1492–1502. https://doi.org/10.1101/gad.282145.116
Hogg A, Bia B, Onadim Z, Cowell J (1993) Molecular mechanisms of oncogenic mutations in tumors from patients with bilateral and unilateral retinoblastoma. Proc Natl Acad Sci USA 90(15):7351–7355. https://doi.org/10.1073/pnas.90.15.7351
Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, Grody WW, Hegde M, Lyon E, Spector E, Voelkerding K, Rehm HL (2015) Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med 17(5):405–424. https://doi.org/10.1038/gim.2015.30
Mohd Khalid MK, Yakob Y, Md Yasin R, Wee Teik K, Siew CG, Rahmat J, Ramasamy S, Alagaratnam J (2015) Spectrum of germ-line RB1 gene mutations in Malaysian patients with retinoblastoma. Mol Vis 21:1185–1190
Parma D, Ferrer M, Luce L, Giliberto F, Szijan I (2017) RB1 gene mutations in Argentine retinoblastoma patients. Implic genetic couns PLoS One 12(12):e0189736. https://doi.org/10.1371/journal.pone.0189736
Cheng G, Wang Y, Bin L, Shi J, Zhao J, Jonas JB (2013) Genetic and epigenetic profile of retinoblastoma in a Chinese population: analysis of 47 patients. Asia Pac J Ophthalmol (Phila) 2(6):414–417. https://doi.org/10.1097/apo.0000000000000016
Abidi O, Knari S, Sefri H, Charif M, Senechal A, Hamel C, Rouba H, Zaghloul K, El Kettani A, Lenaers G, Barakat A (2011) Mutational analysis of the RB1 gene in Moroccan patients with retinoblastoma. Mol Vis 17:3541–3547
Li W, Buckley J, Sanchez-Lara P, Maglinte D, Viduetsky L, Tatarinova T, Aparicio J, Kim J, Au M, Ostrow D, Lee T, O’Gorman M, Judkins A, Cobrinik D, Triche T (2016) A rapid and sensitive next-generation sequencing method to detect RB1 mutations improves care for retinoblastoma patients and their families. J mol diagn: JMD 18(4):480–493. https://doi.org/10.1016/j.jmoldx.2016.02.006
Eloy P, Dehainault C, Sefta M, Aerts I, Doz F, Cassoux N (2016) Lumbroso le Rouic L, Stoppa-Lyonnet D, Radvanyi F, Millot G, Gauthier-Villars M, Houdayer C. a parent-of-origin effect impacts the phenotype in low penetrance retinoblastoma families segregating the c.1981C>T/p.Arg661Trp mutation of RB1. PLoS Genet 12(2):e1005888
Quiñonez-Silva G, Dávalos-Salas M, Recillas-Targa F, Ostrosky-Wegman P, Aranda D, Benítez-Bribiesca L (2016) Monoallelic germline methylation and sequence variant in the promoter of the RB1 gene: a possible constitutive epimutation in hereditary retinoblastoma. Clin Epigenetics 8:1. https://doi.org/10.1186/s13148-015-0167-0
Chen Z, Moran K, Richards-Yutz J, Toorens E, Gerhart D, Ganguly T, Shields CL, Ganguly A (2014) Enhanced sensitivity for detection of low-level germline mosaic RB1 mutations in sporadic retinoblastoma cases using deep semiconductor sequencing. Hum Mutat 35(3):384–391. https://doi.org/10.1002/humu.22488
Dehainault C, Garancher A, Castéra L, Cassoux N, Aerts I, Doz F, Desjardins L, Lumbroso L, de Montes-Oca R, Almouzni G, Stoppa-Lyonnet D, Pouponnot C, Gauthier-Villars M, Houdayer C (2014) The survival gene MED4 explains low penetrance retinoblastoma in patients with large RB1 deletion. Hum Mol Genet 23(19):5243–5250
Rushlow D, Mol B, Kennett J, Yee S, Pajovic S, Thériault B, Prigoda-Lee N, Spencer C, Dimaras H, Corson T, Pang R, Massey C, Godbout R, Jiang Z, Zacksenhaus E, Paton K, Moll A, Houdayer C, Raizis A, Halliday W, Lam W, Boutros P, Lohmann D, Dorsman J, Gallie B (2013) Characterisation of retinoblastomas without RB1 mutations: genomic, gene expression, and clinical studies. Lancet Oncol 14(4):327–334. https://doi.org/10.1016/s1470-2045(13)70045-7
Castéra L, Sabbagh A, Dehainault C, Michaux D, Mansuet-Lupo A, Patillon B, Lamar E, Aerts I, Lumbroso-Le Rouic L, Couturier J, Stoppa-Lyonnet D, Gauthier-Villars M, Houdayer C (2010) MDM2 as a modifier gene in retinoblastoma. J Natl Cancer Inst 102(23):1805–1808. https://doi.org/10.1093/jnci/djq416
Rizzolio F, Lucchetti C, Caligiuri I, Marchesi I, Caputo M, Klein-Szanto AJ, Bagella L, Castronovo M, Giordano A (2012) Retinoblastoma tumor-suppressor protein phosphorylation and inactivation depend on direct interaction with Pin1. Cell Death Differ 19(7):1152–1161. https://doi.org/10.1038/cdd.2011.202
Soliman S, Dimaras H, Khetan V, Gardiner J, Chan H, Héon E, Gallie B (2016) Prenatal versus Postnatal Screening for Familial Retinoblastoma. Ophthalmology 123(12):2610–2617. https://doi.org/10.1016/j.ophtha.2016.08.027
Vogel F (1979) Genetics of retinoblastoma. Hum Genet 52(1):1–54. https://doi.org/10.1007/bf00284597
Funding
This work was supported by the National Natural Science Foundation of China (82072033, 32000391), the 5010 Project of Clinical medicine Research Sun Yat-sen University (2018011), the Natural Science Foundation of Guangdong province (2021A1515010380), Technology Planning Project of Guangdong Province (2020A1414010308), Guangdong Basic and Applied Basic Research Foundation (2019A1515110375), and Guangdong Provincial Medical Science and Technology Research Fund (A2020378). The funding organizations had no role in the design or conduct of this research. The authors declare that no funds, grants, or other support were received during the preparation of this manuscript.
Author information
Authors and Affiliations
Contributions
All authors contributed to the study conception and design. Material preparation was performed by HL, JZ, HG, and PP. Data collection and analysis were performed by BL, RL, LL, and XH. The first draft of the manuscript was written by LL, and all authors commented on previous versions of the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interests
The authors have no relevant financial or nonfinancial interests to disclose.
Ethical approval
The study was approved by the Ethics Committee of The Fifth Affiliated Hospital, Sun Yat-sen University, and was conducted in accordance with the principles of the 1975 Declaration of Helsinki. Written informed consent was obtained from all individual participants included in the study.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Li, L., Li, H., Zhang, J. et al. Five novel RB1 gene mutations and genotype–phenotype correlations in Chinese children with retinoblastoma. Int Ophthalmol 42, 3421–3430 (2022). https://doi.org/10.1007/s10792-022-02341-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10792-022-02341-2