Abstract
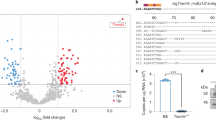
Sepsis-induced tissue and organ damage is caused by an overactive inflammatory response, immune dysfunction, and coagulation dysfunction. Danger-associated molecular pattern (DAMP) molecules play a critical role in the excessive inflammation observed in sepsis. In our previous research, we identified NMI as a new type of DAMP molecule that promotes inflammation in sepsis by binding to toll-like receptor 4 (TLR4) on macrophage surfaces, activating the NF-κB pathway, and releasing pro-inflammatory cytokines. However, it is still unknown whether NMI plays a significant role in other pathways. Our analysis of bulk and single-cell transcriptome data from the GEO database revealed a significant increase in NMI expression in neutrophils and monocytes in sepsis patients. It is likely that NMI functions through multiple receptors in sepsis, including IFNAR1, IFNAR2, TNFR1, TLR3, TLR1, IL9R, IL10RB, and TLR4. Furthermore, the correlation between NMI expression and the activation of NF-κB, MAPK, and JAK pathways, as well as the up-regulation of their downstream pro-inflammatory factors, demonstrates that NMI may exacerbate the inflammatory response through these signaling pathways. Finally, we demonstrated that STAT1 phosphorylation was enhanced in RAW cells upon stimulation with NMI, supporting the activation of JAK signaling pathway by NMI. Collectively, these findings shed new light on the functional mechanism of NMI in sepsis.





Similar content being viewed by others
Availability of Data and Materials
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author. Raw and processed next-generation sequencing datasets are available for download in the NCBI Gene Expression Comprehensive Database (GEO) at numbers: GSE137340, GSE95233 and GSE57065 (bulk RNA-seq); GSE167363 (scRNA-seq).
Abbreviations
- CLRs:
-
C-type lectin receptors
- DAMP:
-
Danger-associated molecular pattern
- DEGs:
-
Differentially expressed genes
- eCIRP:
-
Extracellular cold-inducible RNA-binding protein
- GEO:
-
Gene Expression Omnibus
- GSEA:
-
Gene set enrichment analysis
- GSVA:
-
Gene set variation analysis
- HC:
-
Healthy controls
- HSPs:
-
Heat shock proteins
- HMGB1:
-
High-mobility group box protein 1
- IL-33:
-
Interleukin-33
- NCBI:
-
National Center for Biotechnology Information
- NLRs:
-
NOD-like receptors
- NMI:
-
N-myc and STAT interactor
- PAMPs:
-
Pathogen-associated molecular patterns
- PBMC:
-
Peripheral blood mononuclear cells
- PRRs:
-
Pattern recognizing receptors
- RAGE:
-
Receptor for advanced glycation end products
- RLRs:
-
RIG-like receptors
- scRNA-seq:
-
Single-cell RNA sequencing
- SIRS:
-
Systemic inflammatory response syndrome
- SVA:
-
Surrogate Variable Analysis
- TLR4:
-
Toll-like receptor 4
- TLRs:
-
Toll-like receptors
- TREM-1:
-
Triggering receptor expressed on myeloid cells-1
References
Rudd, K.E., S.C. Johnson, K.M. Agesa, K.A. Shackelford, D. Tsoi, D.R. Kievlan, et al. 2020. Global, regional, and national sepsis incidence and mortality, 1990–2017: Analysis for the Global Burden of Disease Study. Lancet 395 (10219): 200–211.
Stanski, N.L., and H.R. Wong. 2020. Prognostic and predictive enrichment in sepsis. Nature Reviews. Nephrology 16 (1): 20–31.
Shahreyar, M., R. Fahhoum, O. Akinseye, S. Bhandari, G. Dang, and R.N. Khouzam. 2018. Severe sepsis and cardiac arrhythmias. Ann Transl Med. 6 (1): 6.
Ackerman, M.H., T. Ahrens, J. Kelly, and A. Pontillo. 2021. Sepsis. Critical Care Nursing Clinics of North America 33 (4): 407–418.
Matzinger, P. 2002. The danger model: A renewed sense of self. Science 296 (5566): 301–305.
Rubartelli, A., and M.T. Lotze. 2007. Inside, outside, upside down: Damage-associated molecular-pattern molecules (DAMPs) and redox. Trends in Immunology 28 (10): 429–436.
Denning, N.L., M. Aziz, S.D. Gurien, and P. Wang. 2019. DAMPs and NETs in Sepsis. Frontiers in Immunology 10: 2536.
Takeuchi, O., and S. Akira. 2010. Pattern recognition receptors and inflammation. Cell 140 (6): 805–820.
Singer, M., C.S. Deutschman, C.W. Seymour, M. Shankar-Hari, D. Annane, M. Bauer, et al. 2016. The Third International Consensus Definitions for Sepsis and Septic Shock (Sepsis-3). JAMA 315 (8): 801–810.
Salomao, R., B.L. Ferreira, M.C. Salomao, S.S. Santos, L.C.P. Azevedo, and M.K.C. Brunialti. 2019. Sepsis: Evolving concepts and challenges. Brazilian Journal of Medical and Biological Research 52 (4): e8595.
Aziz, M., A. Jacob, W.L. Yang, A. Matsuda, and P. Wang. 2013. Current trends in inflammatory and immunomodulatory mediators in sepsis. Journal of Leukocyte Biology 93 (3): 329–342.
Sunden-Cullberg, J., A. Norrby-Teglund, A. Rouhiainen, H. Rauvala, G. Herman, K.J. Tracey, et al. 2005. Persistent elevation of high mobility group box-1 protein (HMGB1) in patients with severe sepsis and septic shock. Critical Care Medicine 33 (3): 564–573.
Zhang, Q., M. Raoof, Y. Chen, Y. Sumi, T. Sursal, W. Junger, et al. 2010. Circulating mitochondrial DAMPs cause inflammatory responses to injury. Nature 464 (7285): 104–107.
Qiang, X., W.L. Yang, R. Wu, M. Zhou, A. Jacob, W. Dong, et al. 2013. Cold-inducible RNA-binding protein (CIRP) triggers inflammatory responses in hemorrhagic shock and sepsis. Nature Medicine 19 (11): 1489–1495.
Ekaney, M.L., G.P. Otto, M. Sossdorf, C. Sponholz, M. Boehringer, W. Loesche, et al. 2014. Impact of plasma histones in human sepsis and their contribution to cellular injury and inflammation. Critical Care 18 (5).
Denstaedt, S.J., J.L. Spencer-Segal, M.W. Newstead, K. Laborc, A.P. Zhao, A. Hjelmaas, et al. 2018. S100A8/A9 drives neuroinflammatory priming and protects against anxiety-like behavior after sepsis. The Journal of Immunology 200 (9): 3188–3200.
Vulczak, A., C.H.R. Catalao, L.A.P. Freitas, M.J.A. Rocha, 2019. HSP-target of therapeutic agents in sepsis treatment. International Journal of Molecular Science 20 (17).
Nascimento, D.C., P.H. Melo, A.R. Pineros, R.G. Ferreira, D.F. Colon, P.B. Donate, et al. 2017. IL-33 contributes to sepsis-induced long-term immunosuppression by expanding the regulatory T cell population. Nature Communications 8: 14919.
Mouncey, P.R., T.M. Osborn, G.S. Power, D.A. Harrison, M.Z. Sadique, R.D. Grieve, et al. 2015. Trial of early, goal-directed resuscitation for septic shock. New England Journal of Medicine 372 (14): 1301–1311.
Zhou, M., M. Aziz, and P. Wang. 2021. Damage-associated molecular patterns as double-edged swords in sepsis. Antioxidants & Redox Signaling 35 (15): 1308–1323.
Denning, N.L., M. Aziz, A. Murao, S.D. Gurien, M. Ochani, J.M. Prince, et al. 2020. Extracellular CIRP as an endogenous TREM-1 ligand to fuel inflammation in sepsis. JCI Insight 5 (5).
Xiahou, Z., X. Wang, J. Shen, X. Zhu, F. Xu, R. Hu, et al. 2017. NMI and IFP35 serve as proinflammatory DAMPs during cellular infection and injury. Nature Communications 8 (1): 950.
Jing, X., Y. Yao, D. Wu, H. Hong, X. Feng, N. Xu, et al. 2021. IFP35 family proteins promote neuroinflammation and multiple sclerosis. Proceedings National Academy of Sciences USA 118 (32).
Bosmann, M., and P.A. Ward. 2013. The inflammatory response in sepsis. Trends in Immunology 34 (3): 129–136.
Schaefer, L. 2014. Complexity of danger: The diverse nature of damage-associated molecular patterns. Journal of Biological Chemistry 289 (51): 35237–35245.
Song, B., X. Luo, X. Luo, Y. Liu, Z. Niu, X. Zeng, 2022. Learning spatial structures of proteins improves protein-protein interaction prediction. Brief Bioinformatics 23 (2).
Sledzieski, S., R. Singh, L. Cowen, B. Berger. 2021. D-SCRIPT translates genome to phenome with sequence-based, structure-aware, genome-scale predictions of protein-protein interactions. Cell Systems 12 (10):969–82 e6.
Zhang, Q.C., D. Petrey, L. Deng, L. Qiang, Y. Shi, C.A. Thu, et al. 2012. Structure-based prediction of protein-protein interactions on a genome-wide scale. Nature 490 (7421): 556–560.
Kanehisa, M., and S. Goto. 2000. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Research 28 (1): 27–30.
Jin, S., C.F. Guerrero-Juarez, L. Zhang, I. Chang, R. Ramos, C.H. Kuan, et al. 2021. Inference and analysis of cell-cell communication using Cell Chat. Nature Communications 12 (1): 1088.
Janeway, C.A., Jr. 1989. Approaching the asymptote? Evolution and revolution in immunology. Cold Spring Harbor Symposia on Quantitative Biology 54 (Pt 1): 1–13.
Ye, W., X. Liu, Y. Bai, N. Tang, G. Wu, X. Wang, et al. 2021. Sepsis activates the TLR4/MyD88 pathway in Schwann cells to promote infiltration of macrophages, thereby impeding neuromuscular function. Shock 55 (1): 90–99.
Sharma, A., K. Kontodimas, and M. Bosmann. 2021. The MAVS Immune recognition pathway in viral infection and sepsis. Antioxidants & Redox Signaling 35 (16): 1376–1392.
Kolaczkowska, E., and P. Kubes. 2013. Neutrophil recruitment and function in health and inflammation. Nature Reviews Immunology 13 (3): 159–175.
Vogel, S., R. Bodenstein, Q.W. Chen, S. Feil, R. Feil, J. Rheinlaender, et al. 2015. Platelet-derived HMGB1 is a critical mediator of thrombosis. The Journal of Clinical Investigation 125 (12): 4638–4654.
Reyes, M., M.R. Filbin, R.P. Bhattacharyya, K. Billman, T. Eisenhaure, D.T. Hung, et al. 2020. An immune-cell signature of bacterial sepsis. Nature Medicine 26 (3): 333–340.
Clere-Jehl, R., A. Mariotte, F. Meziani, S. Bahram, P. Georgel, and J. Helms. 2020. JAK-STAT targeting offers novel therapeutic opportunities in sepsis. Trends in Molecular Medicine 26 (11): 987–1002.
Tabone, O., M. Mommert, C. Jourdan, E. Cerrato, M. Legrand, A. Lepape, et al. 2018. Endogenous retroviruses transcriptional modulation after severe infection, trauma and burn. Frontiers in Immunology 9: 3091.
Ouyang, W., and A. O’Garra. 2019. IL-10 Family Cytokines IL-10 and IL-22: From Basic Science to Clinical Translation. Immunity 50 (4): 871–891.
Shen, W., Z. Song, X. Zhong, M. Huang, D. Shen, P. Gao, et al. 2022. Sangerbox: A comprehensive, interaction-friendly clinical bioinformatics analysis platform. iMeta 1 (3):e36.
Hänzelmann, S., R. Castelo, and J. Guinney. 2013. GSVA: Gene set variation analysis for microarray and RNA-seq data. BMC Bioinformatics 14: 7.
Satija, R., J.A. Farrell, D. Gennert, A.F. Schier, and A. Regev. 2015. Spatial reconstruction of single-cell gene expression data. Nature Biotechnology. 33 (5): 495–502.
Korsunsky, I., N. Millard, J. Fan, K. Slowikowski, F. Zhang, K. Wei, et al. 2019. Fast, sensitive and accurate integration of single-cell data with Harmony. Nature Methods 16 (12): 1289–1296.
Becht, E., L. McInnes, J. Healy, C.A. Dutertre, I.W.H. Kwok, L.G. Ng, et al. 2018. Dimensionality reduction for visualizing single-cell data using UMAP. National Biotechnology.
Clarke, Z.A., T.S. Andrews, J. Atif, D. Pouyabahar, B.T. Innes, S.A. MacParland, et al. 2021. Tutorial: Guidelines for annotating single-cell transcriptomic maps using automated and manual methods. Nature Protocols 16 (6): 2749–2764.
Aran, D., A.P. Looney, L. Liu, E. Wu, V. Fong, A. Hsu, et al. 2019. Reference-based analysis of lung single-cell sequencing reveals a transitional profibrotic macrophage. Nature Immunology 20 (2): 163–172.
Han, X., Z. Zhou, L. Fei, H. Sun, R. Wang, Y. Chen, et al. 2020. Construction of a human cell landscape at single-cell level. Nature 581 (7808): 303–309.
Singh, R., K. Devkota, S. Sledzieski, B. Berger, and L. Cowen. 2022. Topsy-Turvy: Integrating a global view into sequence-based PPI prediction. Bioinformatics (Oxford, England). 38 (Suppl 1): i264–i272.
Acknowledgements
We thank Prof. Huanhuan Liang (Sun Yat-sen University) for valuable discussion and critical comments on the manuscript. We also thank all the contributors of the public databases used in this study for providing all the data for analysis.
Funding
The work was supported by the grants from Shenzhen Science and technology planning project (project no. JCYJ20200109142412265, ZDSYS20220606100803007 to Y.L., project no. RCBS20200714114922284 to Z.W.; project no. 202206193000001, 20220817122906001 to J.Q.). The funding bodies were not involved in the design of the study, collection, analysis, interpretation of data, or writing of the manuscript.
Author information
Authors and Affiliations
Contributions
J.Z. and Z.Y. collected the data and performed all the analysis. J.Z., Z.Y., and J.S. prepared the images. J.Z., D.X., J.Q., Z.W., and Y.L. wrote the manuscript and organized the study. All authors contributed to the manuscript and approved the version as submitted.
Corresponding authors
Ethics declarations
Ethics Approval and Consent to Participate
Not applicable.
Consent for Publication
Not applicable.
Competing Interests
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Zeng, J., Yang, Z., Xu, D. et al. NMI Functions as Immuno-regulatory Molecule in Sepsis by Regulating Multiple Signaling Pathways. Inflammation 47, 60–73 (2024). https://doi.org/10.1007/s10753-023-01893-4
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10753-023-01893-4