Abstract
Pikeperch (Sander lucioperca Linnaeus, 1758) is a wide-ranged percid predator fish characterised by a great ecological value in the Eurasian freshwater and brackish ecosystems. It is also one of the most famous fish species of Lake Balaton (Hungary), where a unique (genetically separated) pikeperch stock lives. However, until now, no detailed information was available about the pikeperch population genetic structure in Lake Balaton. In the present study, the population genetic structure of the pikeperch assemblages in the lake was revealed by using microsatellite markers. Commercial fishery and angler catch data going back more than 100 years verified that the pikeperch has always been a key element of Lake Balaton’s fish stock utilization. Results of genetic data analyses showed that the pikeperch in the Lake Balaton forms a metapopulation system, in which only the westward stocks show certain separation. Moreover, it seems that the exploitation and mass fish kills that happened in the 1960s and 1970s may have had only a slight impact on the population genetic structure of Balaton pikeperch stocks. The information about genetic features and utilization changes of pikeperch stocks can help to develop area-specific management plans and ensure the long-term survival of this carnivore fish species characteristic of Lake Balaton.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Pikeperch (Sander lucioperca Linnaeus, 1758) is a wide-ranging percid predator fish of the Eurasian freshwater and brackish ecosystems (Hokanson, 1977; Brown et al., 2001; Kottelat & Freyhof, 2007). It is distributed from the southern area of the Balkan Peninsula beyond the Artic Circle in Scandinavia, and the Aral Sea to the River Elbe (Kottelat & Freyhof, 2007). In the centre of its relatively large distribution area, the species is in decline (Olsson, 2019), while at the edge of its distribution, its spread has been detected (Kottelat & Freyhof, 2007; Craig, 2008; Barmintseva et al., 2014; Eschbach et al., 2014; Louati et al., 2016). Since it is one of the top predatory fishes in these above-mentioned habitats, it has great ecological importance, and at the same time, great economic value. It is important to the local fishery in its whole range (Vetamaa et al., 2001; Tyutyunov et al., 2002; Abdolmalaki & Psuty, 2007); similarly, as a favourable game fish, it has considerable recreational use (e.g. Dahl, 1982; Harka & Sallai, 2004; Lehtonen et al., 1996; Steffens & Winkel, 1999). For this reason, this species has been introduced to several watersheds (Eschbach et al., 2014; Louati et al., 2016). Besides traditional exploitation, pikeperch is considered as one of the most promising freshwater fish species for aquaculture in Europe. Its high growth potential, high-quality flesh and high market acceptance make it an appropriate candidate for land-based recirculating aquaculture systems (Dalsgaard et al., 2013; Pyanov et al., 2014; Ende et al., 2021). The demand for good quality pikeperch products has increased, in the case of both natural stocks and aquaculture production. For this reason, the knowledge of ecology, population dynamics and genetics of natural pikeperch stocks can help to develop area-specific management plans for the long-term survival of the species in natural habitats. At the same time, we can get important information for the selection of the most appropriate lineages for aquacultural utilisation.
This information can also help to develop a stock structure more appropriate to increase aquaculture productivity. Since the pikeperch is an important species in fisheries, particularly for anglers, several notes have been published about its genetic features. Results of the macroscale phylogenetic studies of Cytochrome B sequence analyses made on the continental European pikeperch population showed two major clades (Kohlmann et al., 2013). The first clade can be found in Northern Europe and Asia. The second haplogroup is found in Central Europe in the Danube drainage system (Haponski & Stepien, 2013; Tsaparis et al., 2022). The pikeperch is a non-indigenous species in France and Tunisia, but this later haplogroup is still present there. There are several hypotheses regarding how the pikeperch appeared in these areas (Armengaud, 1962; Goubier, 1972), but the microsatellite marker analysis suggests that these populations have Central European origin (Tsaparis et al., 2022). Even though in France one can find only introduced, stocked populations, their allelic richness was higher than was detected at Baltic Sea stocks (Poulet et al., 2009). Smaller-scale studies have revealed that the Finnish lake populations showed greater genetic diversity than the coastal stocks (Säisä et al., 2010). Moreover, a north-to-south genetic diversity gradient was shown in this area (Björklund et al., 2007). Another survey showed that the current stocking practices cause artificial gene flow, which decreases the genetic divergence of natural Finnish pikeperch populations (Salminen et al., 2012). In Asia, there is a significant genetic variation among the pikeperch populations both in the Aral Sea (Khurshut & Kohlmann, 2009), and the Caspian Sea (Gharibkhani et al., 2009). In contrast, in Kazakhstan, there is low genetic divergence between the natural populations, which may be caused by the rapid range expansion of this species (Barmintseva et al., 2014). In Russia, there is no significant difference between the Volga and Akhtuba populations (Kusishchin et al., 2018). In China, instead of the north-to-south genetic separation that characterises the Scandinavian stocks (Björklund et al., 2007; Säisä et al., 2010), a significant east-to-west separation was observed (Lu et al., 2022).
As has been noted above, the Danube system hosts a separate pikeperch lineage. At the same time, the recent study by Tsaparis et al. (2022) highlighted that the Hungarian pikeperch stocks have a unique genetic background and can be characterised by higher genetic diversity compared to the other European populations. Therefore, the pikeperch stocks living in the inner area of the Carpathian basin are treated as a separate cluster. This high-level differentiation can be explained by the fact that the centre of the Carpathian Basin (where Hungary is situated) is mainly lowland, and the environmental circumstances are appropriate for this species. Moreover, this area was never glaciated during the ice ages (Hewitt, 1996); therefore, it could serve as a refugium for many terrestrial and aquatic species (Varga, 2009; Schmitt & Varga, 2012), and a genetically separated cluster of pikeperch could also survive. Since this species prefers shallow eutrophic lakes with low water transparency (Sonesten, 1991; Craig, 2008) Lake Balaton provides appropriate environmental conditions to keep and maintain large pikeperch stocks. Therefore, Lake Balaton has long been recognised as the best and most characteristic pikeperch habitat in Hungary (Harka & Sallai, 2004).
Pikeperch is among the most valuable species in the fishery sector, and it has become one of the most popular targets of anglers in the last decades. To ensure the long-term conservation of the lake’s pikeperch populations, it has been registered with protected designations of origin and protected geographical indications (‘Balatoni hal’ PGI) by the European Commission (“URL1”). Therefore, it is widely accepted that pikeperch is of particular ecological and economic importance in Lake Balaton, and several studies were made on the biology of the species (Bíró, 1981; Specziár, 2010 and the cited works therein). Moreover, little information has been published so far about the genetic variation of Lake Balaton’s pikeperch stocks. Two of the available population genetic surveys (Kánainé Sipos et al., 2019; Molnár et al., 2020) do not provide information about the genetic variation of the pikeperch stock within Lake Balaton.
The Hungarian population has greater genetic diversity than other European populations (Tsaparis et al., 2022). Lake Balaton is the best pikeperch habitat in Hungary (Harka & Sallai, 2004), based on its large size, unique shape and special hydrological characteristics (Istvánovics et al., 2007), as in the case of other large lakes (Egger et al., 2007; Sepulveda-Villet & Stepien, 2011). Our basic hypothesis is that more than one metapopulation may occur in the lake. Therefore, the main aims of this present study are (i) to analyse the genetic structure of the pikeperch stock within the Lake Balaton, which is the largest isolated native population of this species in Central Europe (ii) to reveal any discrepancies within the population and (iii) to evaluate whether the observed patterns are related to any environmental impact.
Materials and methods
Study area
Our population genetic study was carried out on Lake Balaton, which is one of the largest (A: 594 km2, mean depth: 3.2 m, V: ~ 1.8 km3) freshwater shallow lakes in Central Europe (Istvánovics et al., 2007). Lake Balaton is the largest contiguous natural habitat for the pikeperch in the Carpathian Basin. The surface area as well as the mean depth of the basins increase from west to east. The estuaries of the most important inflows are in the western area of the lake. The specific hydrological features and the lake morphometry cause environmental gradients along the longitudinal axis (Istvánovics et al., 2007) from the eu- or mesotrophic western to an oligotrophic eastern area of the lake.
Sample collection
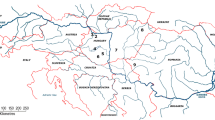
Fin samples were collected from six different locations of Lake Balaton from anglers’ catches during 2016–2017. In all cases, the standard length (SL) of the fish used for the sampling exceeded 350 mm. The cut fin samples were stored in 1.5-mL micro-tubes in 96% ethanol at –20 °C until the start of the DNA isolation. The locations and other important information of samples are shown in Table 1 and Fig. 1.
A The distribution of pikeperch sampling sites on the littoral region of Lake Balaton; geographic position of Hungary in Europe and Lake Balaton in Hungary are indicated in inserts (B) and (C), respectively; the oligo-mesotrophic gradient of the lake is indicated by an arrow; ke- Keszthely population, bo- Balatonboglár population, ba- Balatonakali population, ti- Tihany population, si- Siófok population and bf- Balatonfűzfő population
DNA isolation
Total genomic DNA was extracted using QIAGEN DNeasy Blood & Tissue Kits according to the instructions of the manufacturer. The DNA concentration of the samples was determined with a Maestro NanoDrop spectrophotometer (Thermo Fisher Scientific, Wilmington, Delaware, USA), adjusted to 55 ng/µL for later use, and stored at –20 °C.
PCR amplification and microsatellite analysis
Fifteen microsatellite markers (MSL1, MSL2, MSL3, MSL5, MSL9—Kohlmann & Kersten, 2008; Svi-4, Svi-6, Svi-L7, Svi-18—Wirth et al., 1999; Pfla-L3, Pfla-L9—Leclerc et al., 2000; Za038, Za144, Za199, Za207, Za237—Dubut et al., 2010) were used to genotype the individuals. In the PCR reaction with NED, PET, VIC, and FAM end-labelled primers, the amplification of markers was performed in three multiplexes and one simplex reaction in a reaction volume of 20 µL. The composition of the multiplex A reaction was 1.5 mM MgCl2, 200 μM dNTP, 0.1 μM MSL1, 0.066 μM MSL3, 0.266 μM MSL5, 0.2 μM MSL9 from primer, 55 ng template DNA and 1.2 U Taq polymerase. The composition of the multiplex B reaction was 1.5 mM MgCl2, 200 μM dNTP, 0.2 μM MSL2, 0.1 μM Svi-4, 0.1 μM Svi-6, 0.2 μM Svi-L7, 0.2 μM from Svi-18, 0.2 μM from Pfla-L8 primer, 55 ng of template DNA and 1.2 U of Taq polymerase. The composition of the third multiplex reaction was 1.5 mM MgCl2, 200 μM dNTP, 0.06 μM PflaL9, 0.02 μM Za038, 0.04 μM Za144, 0.06 μM Za207, 0.06 μM from Za237 primer, 55 ng of template DNA and 1.2 U of Taq polymerase. The composition of the simplex reaction was 1.5 mM MgCl2, 200 μM dNTP, 0.01 μM Za199 primer, 55 ng template DNA and 1.2U Taq polymerase.
The temperature profile of the PCR reactions was as follows: multiplex A and B: pre-denaturation at 94 °C for ten minutes, thirty-five cycles: 60 s at 94 °C, 90 s at the coupling temperature (multiplex A: 56 °C, multiplex B: 55 °C), and 60 s at 72 °C, and then the PCR product was kept at 4 °C. For multiplex C, the profile was pre-denaturation at 94 °C, two cycles, 60 s at 94 °C, 90 s at 59 °C and 60 s at 72 °C, two cycles, 60 s at 94 °C, 90 s at 58 °C and 60 s at 72 °C, two cycles, 60 s at 94 °C, 90 s at 56 °C and 60 s at 72 °C, 25 cycles, 60 s at 94 °C, 90 s at 59 °C and 60 s at 72 °C and then the PCR product was kept at 4 °C. The simplex PCR temperature profile was the following: pre-denaturation at 94°C, five cycles, 60 s at 94 °C, 90 s at 49 °C and 60 s at 72 °C, thirty cycles, 60 s at 94 °C, 90 s at 47 °C and 60 s at 72 °C, and then the PCR product was kept at 4 °C. The length of the PCR products was examined on an eight-capillary ABI 3500 type sequencer (POP-7 polymer, GeneScan standard 600 LIZ). The fragment sizes were quantified using GeneMapper 4.1 software.
Processing genetic data
MICRO-CHECKER version 2.2.3 (number of randomisations: 1000, 95% CI) was used to evaluate the presence of null alleles (Van Oosterhout et al., 2006). The number of alleles (Na), number of effective alleles (Neff), observed (Ho) and unbiased expected heterozygosity (uHe), and fixation index (F) were calculated using the GenAlEx 6.5 programme (Peakall & Smouse, 2012). Allele richness (AR) and individual allele richness (ARp) were estimated by HP RARE 1.0 (Kalinowski, 2005). The comparison of genetic diversity values in each subpopulation was performed by one-way ANOVA (Tukey post hoc test, in case of Na, AR, Ho, uHe and F) and Kruskal–Walli’s test with Bonferroni correction (a significance threshold of 0.008, in case of Neff and ARp) using the SPSS 11.5.0 software package. The analysis of the molecular variance (AMOVA), after which genetic differentiation among the population (Fst) values were determined in all subpopulation pairs, the calculation of the Nei’s genetic distance matrix of the individuals was performed using the GenAlEx 6.5 programme. The Neighbor Joining (NJ) tree was constructed based on Nei’s genetic distances using MEGA11 software (Tamura et al., 2021). The Bayesian algorithm implemented in the software STRUCTURE (Pritchard et al., 2000; Falush et al., 2003) was used to determine population structure. The most probable cluster number (K) was estimated by using both posterior probabilities (highest lnP(D)) and the ΔK method of Evanno et al. (2005) in the STRUCTURE HARVESTER software (Earl & vonHoldt, 2012). To determine the cluster number, an admixture scenario with allele frequencies correlated was chosen, the burn-in was set to 10,000, and the number of further MCMC runs was set to 200,000. Calculations were repeated 10 times for each K. Discriminant analysis of principal components (DAPC) using microsatellite loci and populations was performed in the R environment (4.2.1) with the adegenet 2.1.1.7 package (Jombart, 2008). The paired Mantel test (9999 permutations), between pairwise Fst values and geographic distances among populations, was calculated in the GenAlEx 6.5 software. Genetic bottleneck, indicating potential population declines, was tested with Bottleneck 1.2.02 software (Cornuet & Luikart, 1996) under a two-phased mutation model (TPM) with 0% stepwise mutation model (SMM) in the TPM and 36% variance of the geometric distribution. The significance was estimated by the Wilcoxon sign-rank test with 1000 iterations. The effective population size (Ne) was estimated with LD and heterozygote excess methods implemented in NeEstimator 2.01 software (Do et al., 2014). The relative directional migration network was calculated by divMigrate-online software (Sundqvist et al., 2016) using the method based on Jost's D and 1000 bootstrap iterations for the statistical testing of the asymmetry between migration rates of all population pairs. A threshold value of 0.5 was used in the filter function to highlight the main migration directions.
Results
Genetic diversity of the populations
The Microchecker did not detect evidence for large allelic dropout, and the presence of null alleles was assumed only in the case of loci PflaL9 (in the Tihany population) and MSL-5 (in the Balatonfűzfő population) due to a general excess of homozygotes. The genetic diversity data of the six populations are shown in Table 2. The number of alleles (Na) was significantly higher in the Tihany population compared to the other populations except for Keszthely. However, considering the effective number of alleles, the Keszthely population showed a significantly higher value. The same tendency was detected in allelic richness (AR) and private allelic richness (ARp), where the value in Keszthely was the highest, but the difference was significant only for ARp. Both unbiased expected heterozygosity (uHe) and observed heterozygosity (Ho) were significantly higher in the Keszthely population. The fixation index had a low negative value in all populations except Balatonfűzfő, and the deviation from Hardy–Weinberg equilibrium was significant at the Za237 locus in the Balatonboglár and Balatonakali populations, the MSL-5, MSL-9 and PfLa-L9 in the Tihany population, Svi-4 in the Siófok population and Svi-6, MSL-2 and MSL-5 in the Balatonfűzfő population.
Genetic structure of the pikeperch stock in the Lake Balaton
The AMOVA revealed in a low level of genetic differentiation among populations (Fst = 0.028, P < 0.001); the most genetic variance was detected within individuals (92%, P < 0.001) and the proportion among individuals was 5% (P < 0.001). However, the pairwise comparisons showed higher differentiation between the population pairs (Table 3.) The Keszthely population showed a significant differentiation from all the other populations based on the Fst values (Table 3). Moreover, the Keszthely population also showed higher Fst values than 0.05 with the Tihany, Siófok and Balatonfűzfő populations.
The genetic separation of the Keszthely population was also supported by the NJ tree constructed based on Nei’s genetic distance (Fig. 2A). Bayesian analysis with the STRUCTURE program resulted in the most probable cluster number K = 2 according to the lnP(D) values (Fig. 2B). However, based on the Evanno et al. (2005) method, the change in ΔK value was the highest in the case of five clusters within the pikeperch stock in Lake Balaton (Fig. 2B). The STRUCTURE results show no distinct clusters, neither for K = 2 nor for K = 5. This reflects the fact that the Fst values between populations were found to be low and thus not high enough for STRUCTURE to identify distinct clusters. Neither the two nor the five clusters determined by the software were associated with the distinct populations.
Genetic structure of the pikeperch stock in the Lake Balaton; A shows the NJ tree based on the Nei’s genetic distances; B represents bar plots resulting from STRUCTURE analysis for K = 2 and K = 5 values. ke-Keszthely population, bo- Balatonboglár population, ba- Balatonakali population, ti- Tihany population, si- Siófok population and bf- Balatonfűzfő population
As a result of the DAPC analysis, the Keszthely population also shows separation (Fig. 3A). The Keszthely population also shows a clear membership probability for almost all the individuals (Fig. 3B) in contrast to the mixed pattern of the other populations.
Genetic structure of the pikeperch stock in Lake Balaton based on DAPC analysis; part A: scatterplot for DAPC analysis of the six populations; part B: membership probabilities of the individuals based on the DAPC analysis; ke-Keszthely population, bo- Balatonboglár population, ba- Balatonakali population, ti- Tihany population, si- Siófok population and bf- Balatonfűzfő population
Gene flow, population size, and bottleneck in the pikeperch stock of Lake Balaton
The Mantel test demonstrated a strong correlation between genetic (Fst) and geographical distance (Fig. 4). Based on the regression, a geographical distance of 42 km is required to reach the Fst value of 5%, which already shows a meaningful differentiation.
The effective population sizes estimated by the Ht and Ld method and the occurrence of the genetic bottleneck events are shown in Table 4. The lowest estimated effective population size was in the Balatonboglár population, which showed shifted allele frequency distribution, indicating a recent bottleneck event in the population. In the Tihany population, significant heterozygosity deficiency was detected, but the allele frequency showed a normal L-shaped distribution. However, the Keszthely population showed significant heterozygosity excess.
The calculated relative directional migration matrix (Fig. 5A.) showed the lowest migration from Keszthely to Siófok (0.063) and the highest value from Siófok to Tihany (1.000). The partial separation of the Keszthely population was easily detected if the filter threshold was set to 0.5; the Tihany population showed the strongest two-way migration with all other populations (except for Keszthely) (Fig. 5B). The bootstrapping confirmed that statistically significant asymmetrical migration occurs in all cases relative to the Keszthely population (Fig. 5C).
Directional relative migration estimated by divMigrate-online (D method): A all populations, no filter was used, B all populations, the filter threshold was set to 0.5; C asymmetric migration, 1000 bootstrapping, no filter was used; ke- Keszthely population, bo- Balatonboglár population, ba- Balatonakali population, ti- Tihany population, si- Siófok population and bf- Balatonfűzfő population
Discussion
Genetic diversity and utilisation changes
The genetic diversity (He: 0.55–0.74, Ar: 4.17–5.42) of the pikeperch populations in Lake Balaton is highly comparable to the literature data. In Finnish populations: in coastal populations, He = 0.30–0.37, Ar = 3.3–3.9, in lake populations He = 0.36–0.46, Ar = 4.0–5.5 (Säisä et al., 2010) or He = 0.51, Ar = 3.6–4.6 (Salminen et al., 2012). In the Rhone delta, He = 0.64–0.74 and Ar = 4.0–6.0 (Poulet et al., 2009), and in the lower Volga Basin, He = 0.79–0.82 and Ar = 9.57–10.64 (Kusishchin et al., 2018). Lake Balaton belongs to the Danube River basin, in which the German populations showed lower diversity values (He: 0.62 and Ar: 4.5) (Eschbach et al., 2014). Compared to data in a study covering European stocks and showing a high overlap in the markers used (Tsaparis et al., 2022) with this study, the diversity of the Balaton pikeperch population was also high, not only in the wild (He: 0.40–0. 69, Ar: 2.7–4.9) but also in domesticated stocks (He: 0.35–0. 72, Ar 2.6–5.5); where the stocks with high diversity were of Lake Balaton or Hungarian origin.
Although most subpopulations showed no signs of a genetic bottleneck event (i.e. temporal population decline(s) in the past), the results are not entirely conclusive. In the case of Tihany, several markers showed deviations from HW equilibrium and significant heterozygosity deficiency was detected, but the allele frequency showed a normal L-shaped distribution. It can be assumed that at least for this subpopulation there has been a major recent change in the population size. The Lake Balaton’s fisheries statistics (the catch statistics of the Balaton Fish Management Non-Profit Ltd (and of the legal predecessor companies) and the National Fishery Data Repository), providing information about the period from 1901 to 2020 (Fig. S1), show that the total pikeperch catch in this period was 10,022 tonnes, and an average (± SD) of 83.51 (± 50.18) tonnes of pikeperch were caught yearly. The largest commercial catch was 236.7 tonnes in 1934, while only 16 tons of pikeperch were caught in 2008. Catch data shows considerable temporal changes in population exploitation. In the first decade of the twentieth century, the catch was about 100 tonnes/year, which was followed by a considerable decrease. From the beginning of the 1920s decade, there was an increasing trend, which was followed by a decrease till the middle of the 1940s. The decline due to the Second World War was followed by a fast run-up to the mid-1960s, when the pikeperch catch was around 160–170 tonnes per year. From this period, the commercial pikeperch catches steadily reduced to a few tonnes per year until 2011, when recreational angling was given priority, and for this reason, since 2013, commercial fishing has been prohibited. In the last decade, the anglers’ catch has grown, and the total amount of pikeperch caught from Lake Balaton exceeded 92 tonnes in 2019, which is 36.6% of the total catch of 250.8 tonnes of pikeperch caught from natural waters in Hungary. The recent exploitation rate of the pikeperch population by angling is considerable and is estimated to be 49–56%/year for legal size classes (> 35 cm standard length) based on tagging experiments (Specziár, 2010; Specziár & Turcsányi, 2017). The fact that only one analysed pikeperch population showed traces of bottleneck suggests that neither the harvesting and stocking nor the mass fish kills of the last century (Bíró, 1997) considerably affected the population genetic conditions of Lake Balaton’s pikeperch stock. This may be due to the relatively large size of the pikeperch stock (note the area of the lake is about 600 km2) and the unhindered admixture of the pikeperch stocks.
Population genetic features of pikeperch stocks
Results of the population genetic survey showed weak separation among the studied stocks. The pikeperch stocks are likely to show a metapopulation structure. The existence of the metapopulation may also support the significant, but relatively low pairwise Fst values compared to the other Hungarian and Baltic populations. (Björklund et al., 2007; Säisä et al., 2010; Kánainé Sipos et al., 2019). Spawning migration rarely exceeds 35 km in pikeperch (Lappalainen et al., 2003), while the length of Lake Balaton is 79 km. Therefore, the shape and length of the lake already allow for a minimal degree of isolation by distance, with Fst values greater than 0.05 above 50 km geographic distance showing a minimal degree of genetic isolation. However, distinguishing locally reproducing populations and mapping fine population structure is often difficult, even when using genetic markers. A good example of this is North America's Lake Erie, where fine-scale studies have been conducted on several percid species for such purposes. In the case of yellow perch, although individual spawning groups were well separated genetically, genetic isolation based on geographic distance was not observed, and the genetic effect of some groups on the overall population was greater than others (Sepulveda-Villet & Stepien, 2011). Several studies have been conducted on the North American relative of pikeperch, the walleye. Although Stepien et al. (2012), using microsatellite makers, detected consistent genetic structure in the lake walleye stock by the isolation of one spawning site (Van Buren Bay), other spawning groups showed greater similarity in some years, and self-assignment tests did not show high values in many groups. In a more recent study using RAD sequencing (Chen et al., 2020) for Lake Erie walleye, a more accurate picture was obtained, showing that the western spawning groups showed low separation, but the western and eastern basins were more strongly separated, with high 95% classification accuracy. Our present study showed a similar genetic pattern in the Lake Balaton pikeperch population, and it would be worthwhile to compare subpopulations using SNP markers.
Additionally, the migration computations showed a one-way east-to-west direction for pikeperch movement. This result is in accordance with the findings of Specziár and Turcsányi (2017), who analysed the movement of marked pikeperch individuals. Fish stocked in mesotrophic areas travel smaller distances and disperse less than those stocked in the oligotrophic areas (Specziár & Turcsányi, 2017). It seems, therefore, that the movement and distribution of the pikeperch are strongly related to the trophic gradient of Lake Balaton (Istvánovics 2007). The separation of the stock of Keszthely can also be explained by the different mean depth and surface area, the specific morphometrics of the lake, and the differences between the compositions of the fish fauna (Bíró, 1997). These factors probably resulted in local adaptations within the stock, which would be useful information for stock replacement plantations.
Finally, the genetic structure may also reflect the effects of stocking and introgression (which could be more pronounced in the central and eastern areas). All the pikeperch stocked in Lake Balaton had a natural origin by hatching from artificial nests placed in the lake. Although we do not have information on the local variation in the proportion of introductions for all periods, it could theoretically not affect the genetic structure if the natural population size is large enough. However, there was a significant decline in the pikeperch population in Lake Balaton between 1965 and 1975 due to mass mortalities. At that time, the population size was estimated to have been reduced to between half and one-sixth depending on the area. In the north-eastern part, a survey carried out in 1978 showed a lack of older age classes (the population consisted of 3–5-year fish), which changed towards the south-west, with a dominance of 4–6-year classes in the central part, while in the south-western part (Keszthely), 4–8-year fish were prevalent (Bíró, 1981). Stocking on a reduced natural spawning stock could theoretically have caused introgression (Molnár et al., 2020) in the central and mainly eastern basins. Although, it is not possible to make firm conclusions without historical samples and temporal genetic analyses, anthropogenic influences may have been involved in the development of the present structure.
Conclusions
Commercial fishery and angler catch data going back more than 100 years verified that the pikeperch has always been a key element of Lake Balaton’s fish stock utilisation. Despite the strong recovery of the stocks, the genetic diversity of the pikeperch in Lake Balaton is sufficient compared to other natural stocks. Neither the harvesting and stocking nor the mass fish kills in the 1960s and 1970s appear to have caused bottleneck effects on the species’ genetic pattern. A metapopulation system can be observed in the studied area, in which only the westward stocks (in the Keszthely Bay) show certain separation. The migration calculations showed a one-way east–west direction of pikeperch movement, which is closely related to the trophic gradient observed in Lake Balaton.
Data availability
All data are available in the text or in the supplementary material.
References
Abdolmalaki, S. & I. Psuty, 2007. The effects of stock enhancement of pikeperch (Sander lucioperca) in Iranian coastal waters of the Caspian Sea. ICES Journal of Marine Science 64: 973–980.
Armengaud, J., 1962. Contribution al’étude de Sander lucioperca L, Université Montpellier II, Montpellier, DESS:
Barmintseva, A. E., G. M. Shalgimbayeva, S. K. Koishybayeva, S. T. Sarbakanova, S. Z. Asylbekova, K. B. Isbekov & N. S. Mugue, 2014. Genetic study of pike perch Sander lucioperca (Linnaeus, 1758) and bersh Sander volgensis (Gmelin, 1789) from the fishery water-bodies of Kazakhstan. Russian Journal of Genetics 50: 749–756.
Bíró, P., 1997. Temporal variation in Lake Balaton and its fish populations. Ecology of Freshwater Fish 6: 196–216.
Bíró, P., 1981. A Balaton Halállományának struktúrális változásai (structural changes in fish stocks of Lake Balaton): A Balaton Kutatás Újabb Eredményei II—VEAB Monográfia 16., Veszprém, pp. 239–275. (In Hungarian with English summary).
Björklund, M., T. Aho & L. C. Larsson, 2007. Genetic differentiation in pikeperch (Sander lucioperca): the relative importance of gene flow, drift and common history. Journal of Fish Biology 71: 264–278.
Brown, J. A., W. M. Moore & E. S. Quabius, 2001. Physiological effects of saline waters on Zander. Journal of Fish Biology 59: 1544–1555.
Chen, K. Y., P. T. Euclide, S. A. Ludsin, W. A. Larson, M. G. Sovic, H. L. Gibbs & E. A. Marschall, 2020. RAD-seq refines previous estimates of genetic structure in Lake Erie walleye. Transactions of the American Fisheries Society 149: 159–173.
Cornuet, J. M. & G. Luikart, 1996. Description and power analysis of two tests for detecting recent population bottlenecks from allele frequency data. Genetics 144: 2001–2014.
Craig, J. F., 2008. Percid fishes: systematics, ecology and exploitation, John Wiley & Sons:
Dahl, J., 1982. A century of pikeperch in Denmark. Food and Agriculture Organization of the United Nations, Rome, pp. 344–352. https://www.fao.org/3/ae997b/ae997b.pdf#page=75
Dalsgaard, J., I. Lund, R. Thorarinsdottir, A. Drengstig, K. Arvonen & P. B. Pedersen, 2013. Farming different species in RAS in Nordic countries: current status and future perspectives. Aquacultural Engineering. 53: 2–13. https://doi.org/10.1016/j.aquaeng.2012.11.008.
Do, C., R. S. Waples, D. Peel, G. M. Macbeth, B. J. Tillett & J. R. Ovenden, 2014. NeEstimator v2: re-implementation of software for the estimation of contemporary effective population size (Ne) from genetic data. Molecular Ecology Resources 14: 209–214.
Dubut, V., R. Grenier, E. Meglécz, R. Chappaz, C. Costedoat, D. Danancher, S. Descloux, T. Malausa, J. F. Martin, N. Pech & A. Gilles, 2010. Development of 55 novel polymorphic microsatellite loci for the critically endangered Zingel asper L. (Actinopterygii: Perciformes: Percidae) and cross-species amplification in five other percids. European Journal of Wildlife Research 56: 931–938.
Earl, D. A. & B. M. vonHoldt, 2012. Structure harvester: a website and program for visualizing structure output and implementing the Evanno method. Conservation Genetics Resources 4: 359–361.
Egger, B., S. Koblmüller, C. Sturmbauer & K. M. Sefc, 2007. Nuclear and mitochondrial data reveal different evolutionary processes in the Lake Tanganyika cichlid genus Tropheus. BMC Evolutionary Biology 7: 1–14.
Ende, S. S. W., E. Larceva, M. Bögner, V. Lugert, M. J. Slater & J. Henjes, 2021. Low turbidity in recirculating aquaculture systems (RAS) reduces feeding behavior and increases stress-related physiological parameters in pikeperch ( Sander lucioperca ) during grow-out. Translational Animal Science. https://doi.org/10.1093/tas/txab223.
Eschbach, E., A. W. Nolte, K. Kohlmann, P. Kersten, J. Kail & R. Arlinghaus, 2014. Population differentiation of zander (Sander lucioperca) across native and newly colonized ranges suggests increasing admixture in the course of an invasion. Evolutionary Applications 7: 555–568.
Evanno, G., S. Regnaut & J. Goudet, 2005. Detecting the number of clusters of individuals using the software structure: a simulation study. Molecular Ecology 14: 2611–2620. https://doi.org/10.1111/j.1365-294X.2005.02553.x.
Falush, D., M. Stephens & J. Pritchard, 2003. Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics 164: 1567–1587.
Gharibkhani, M., M. Pourkazemi, M. Soltani, S. Rezvani & L. Azizzadeh, 2009. Population genetic structure of pikeperch (Sander lucioperca Linnaeus, 1758) in the southwest Caspian Sea using microsatellite markers. Journal of Fisheries and Aquatic Science 4: 161–168.
Goubier, J., 1972. Acclimatation du Sandre (Sander lucioperca L.) dans les eaux françaises: Avec 3 figures et 2 tableaux dans le texte. Internationale Vereinigung Für Theoretische Und Angewandte Limnologie: Verhandlungen 18: 1147–1154.
Haponski, A. E. & C. A. Stepien, 2013. Phylogenetic and biogeographical relationships of the Sander pikeperches (Percidae: Perciformes): patterns across North America and Eurasia. Biological Journal of the Linnean Society 110: 156–179.
Harka, Á., & Z. Sallai, 2004. Magyarország halfaunája. Nimfea Természetvédelmi Egyesület, Szarvas, 269. https://scholar.google.com/scholar?hl=hu&as_sdt=0%2C5&q=Harka+Sallai+2004&btnG=
Hewitt, G. M., 1996. Some genetic consequences of ice ages, and their role in divergence and speciation. Biological Journal of the Linnean Society 58: 247–276.
Hokanson, K. E., 1977. Temperature requirements of some percids and adaptations to the seasonal temperature cycle. Journal of the Fisheries Research Board of Canada 34: 1524–1550.
Istvánovics, V., A. Clement, L. Somlyódy, A. Specziár, L. G. Tóth & J. Padisák, 2007. Updating water quality targets for shallow Lake Balaton (Hungary), recovering from eutrophication. Hydrobiologia 581: 305–318.
Jombart, T., 2008. Adegenet: a R package for the multivariate analysis of genetic markers. Bioinformatics 24: 1403–1405.
Kalinowski, S. T., 2005. hp-rare 1.0: a computer program for performing rarefaction on measures of allelic richness. Molecular Ecology 5: 187–189. https://doi.org/10.1111/j.1471-8286.2004.00845.x.
Kánainé Sipos, D., G. Kovács, E. Buza, K. Csenki-Bakos, Á. Ősz, U. Ljubobratović, R. Cserveni-Szücs, M. Bercsényi, I. Lehoczky, B. Urbányi & B. Kovács, 2019. Comparative genetic analysis of natural and farmed populations of pike-perch (Sander lucioperca). Aquaculture International 27: 991–1007.
Khurshut, E. & K. Kohlmann, 2009. Application of nine species-specific microsatellite loci to characterize three pike-perch (Sander lucioperca) populations from the Aral Sea basin in Uzbekistan. Environmental Biotechnology 5: 3–10.
Kohlmann, K. & P. Kersten, 2008. Isolation and characterization of nine microsatellite loci from the pike-perch, Sander lucioperca (Linnaeus, 1758). Molecular Ecology Resources 8: 1085–1087.
Kohlmann, K., M. Louati, P. Kersten, L. Bahri-Sfar, N. Poulet & O. K. Ben Hassine, 2013. Detection of two major cytochrome b lineages in pike-perch, Sander lucioperca, and first data on their distribution in European populations. Environmental Biotechnology 9: 1–5.
Kottelat, M., & J. Freyhof, 2007. Handbook of European freshwater fishes. http://aflimno.free.fr/fichiers/AIDEGESTION/Kottelat.pdf.
Kusishchin, K. V., E. V. Ponomareva, K. Y. Samoilov, M. A. Gruzdeva, M. V. Kholodova & D. S. Pavlov, 2018. Morphological and genetic traits of pikeperch Sander lucioperca of the Volga-Akhtuba aquatic system: on the spatial structure of a species in the lower Volga Basin. Journal of Ichthyology 58: 318–332.
Lappalainen, J., H. Dorner & K. Wysujack, 2003. Reproduction biology of pikeperch (Sander lucioperca (L.))—a review. Ecology of Freshwater Fish 12: 95–106.
Leclerc, D., T. Wirth & L. Bernatchez, 2000. Isolation and characterization of microsatellite loci in the yellow perch (Perca flavescens), and cross-species amplification within the family Percidae. Molecular Ecology 9: 995–997.
Lehtonen, H., S. Hansson, & H. Winkler, 1996. Biology and exploitation of pikeperch, Stizostedion lucioperca (L.), in the Baltic Sea area.In Annales Zoologici Fennici (pp. 525–535). Finnish Zoological and Botanical Publishing Board.
Louati, M., K. Kohlmann, O. K. Ben Hassine, P. Kersten, N. Poulet & L. Bahri-Sfar, 2016. Genetic characterization of introduced Tunisian and French populations of pike-perch (Sander lucioperca) by species-specific microsatellites and mitochondrial haplotypes. Czech Journal of Animal Science 61: 159–171.
Lu, C., Z. Sun, P. Xu, R. Na, W. Lv, D. Cao, T. Liu & X. Zheng, 2022. Novel microsatellites reveal wild populations genetic variance in pike-perch (Sander lucioperca) in China. Aquaculture Reports 23: 101031.
Molnár, T., I. Benedek, B. Kovács, A. Zsolnai & I. Lehoczky, 2020. Genetic consequences of pond production of a pikeperch (Sander lucioperca L) stock with natural origin: the effects of changed selection pressure and reduced population size. PeerJ 8: 8745.
Olsson, J., 2019. Past and current trends of coastal predatory fish in the Baltic Sea with a focus on perch, pike, and pikeperch. Fishes 4: 7.
Peakall, R. & P. E. Smouse, 2012. GenALEx 6.5: genetic analysis in excel: population genetic software for teaching and research-an update. Bioinformatics 28: 2537–2539.
Poulet, N., P. Balaresque, T. Aho & M. Björklund, 2009. Genetic structure and dynamics of a small introduced population: the pikeperch, Sander lucioperca, in the Rhône delta. Genetica 135: 77–86.
Pritchard, J., M. Stephens & P. Donnelly, 2000. Inference of population structure using multilocus genotype data. Genetics 155: 945–959.
Pyanov, D., A. Delmukhametov, & E. Khrustalev, 2014. Pike-perch farming in recirculating aquaculture systems (RAS) in the Kaliningrad region: pike-perch farming in recirculating aquaculture systems (RAS) in the Kaliningrad region. In 9th Baltic Conference on Food Science and Technology “Food for Consumer Wel”.
Säisä, M., M. Salminen, M. L. Koljonen & J. Ruuhijärvi, 2010. Coastal and freshwater pikeperch (Sander lucioperca) populations differ genetically in the Baltic Sea basin. Hereditas 147: 205–214.
Salminen, M., M. L. Koljonen, M. Säisä & J. Ruuhijärvi, 2012. Genetic effects of supportive stockings on native pikeperch populations in boreal lakes—three cases, three different outcomes. Hereditas 149: 1–15.
Schmitt, T. & Z. Varga, 2012. Extra-Mediterranean refugia: the rule and not the exception? Frontiers in Zoology 9: 1–12.
Sepulveda-Villet, O. J. & C. A. Stepien, 2011. Fine-scale population genetic structure of the yellow perch Perca flavescens in Lake Erie. Canadian Journal of Fisheries and Aquatic Sciences 68: 1435–1453.
Sonesten, L., 1991. The biology of pikeperch-a literature review: information fraan Soetvattenslaboratoriet, Drottningholm (Sweden) (In Swedish with English summary).
Specziár, A., 2010. A Balaton halfaunája: a halállomány összetétele, az egyes halfajok életkörülményei és a halállomány korszerű hasznosításának feltételrendszere. Acta Biol. Debr. Oecol. Hung. 23: 7–185.
Specziár, A. & B. Turcsányi, 2017. Management of pikeperch stocking in Lake Balaton: effect of season, area, fish size and method of release on the rate and distribution of recaptures. Knowledge & Management of Aquatic Ecosystems 418: 52.
Steffens, W. & M. Winkel, 1999. Current status and socio-economic aspects of recreational fisheries in Germany. Fisheries Centre Research Reports. 7: 130–133.
Stepien, C. A., J. A. Banda, D. M. Murphy & A. E. Haponski, 2012. Temporal and spatial genetic consistency of walleye spawning groups. Transactions of the American Fisheries Society 141: 660–672.
Sundqvist, L., K. Keenan, M. Zackrisson, P. Prodöhl & D. Kleinhans, 2016. Directional genetic differentiation and relative migration. Ecology and Evolution 6: 3461–3475.
Tamura, K., G. Stecher & S. Kumar, 2021. MEGA11: molecular evolutionary genetics analysis version 11. Molecular Biology and Evolution 38: 3022–3027.
Tsaparis, D., T. Lecocq, D. Kyriakis, K. Oikonomaki, P. Fontaine & C. S. Tsigenopoulos, 2022. Assessing genetic variation in wild and domesticated pikeperch populations: implications for conservation and fish farming. Animals 12: 1178.
Tyutyunov, Y., I. Senina, C. Jost & R. Arditi, 2002. Risk assessment of the harvested pike-perch population of the Azov Sea. Ecological Modelling 149: 297–311.
URL1 (n.d.). https://www.hirbalaton.hu/itt-kaphato-majd-a-balatoni-hal-a-magas-minosegu-eredetmegjelolessel-vedett-termek/
Van Oosterhout, C., D. Weetman & W. F. Hutchinson, 2006. Estimation and adjustment of microsatellite null alleles in nonequilibrium populations. Molecular Ecology Notes 6: 255–256.
Varga, Z., 2009. Extra-Mediterranean refugia, post-glacial vegetation history and area dynamics in Eastern Central Europe. In Relict species (pp. 57–87). Springer, Berlin, Heidelberg. https://link.springer.com/chapter/https://doi.org/10.1007/978-3-540-92160-8_3
Vetamaa, M., V. Vanio, T. Saat & S. Kuldin, 2001. Co-operative fisheries management of the cross border Lake Peipsi-Pihkva. Fisheries Management and Ecology 8: 443–451.
Wirth, T., R. Saint-Laurent & L. Bernatchez, 1999. Isolation and characterization of microsatellite loci in the walleye (Stizostedion vitreum), and cross-species amplification within the family Percidae. Molecular Ecology 8: 1960–1962.
Acknowledgements
The research presented in the article was carried out within the framework of the Széchenyi Plan Plus programme with the support of the RRF 2.3.1 21 2022 00008 project.
Funding
Open access funding provided by Hungarian University of Agriculture and Life Sciences. The research was funded by the Sustainable Development and Technologies National Programme of the Hungarian Academy of Sciences (FFT NP FTA, NP2022-II3/2022). Péter Takács supported by the OTKA FK 140902 project.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interest
The authors declare that they have no competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Additional information
Handling editor: Christian Sturmbauer
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
10750_2023_5347_MOESM4_ESM.tif
Supplementary file4 (TIF 513 KB) Fig. S1 The yearly pikeperch catches in Lake Balaton in the last 120 years; data on commercial fishery (blue line) are provided by the Balaton Fish Management Non-Profit Ltd (and of the legal predecessor companies), while the recreational fishery data (red line) are from the National Fishery Data Repository
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Bánó, B., Benedek, I., Zsolnai, A. et al. Population genetic structure of intensively exploited pikeperch (Sander lucioperca) in Lake Balaton (Hungary). Hydrobiologia 851, 559–571 (2024). https://doi.org/10.1007/s10750-023-05347-8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10750-023-05347-8