Abstract
Lotic freshwater macroinvertebrate species distribution models (SDMs) have been shown to improve when hydrological variables are included. However, most studies to date only include data describing climate or stream flow-related surrogates. We assessed the relative influence of climatic and hydrological predictor variables on the modelled distribution of macroinvertebrates, expecting model performance to improve when hydrological variables are included. We calibrated five SDMs using combinations of bioclimatic (bC), hydrological (H) and hydroclimatic (hC) predictor datasets and compared model performance as well as variance partition of all combinations. We investigated the difference in trait composition of communities that responded better to either bC or H configurations. The dataset bC had the most influence in terms of proportional variance, however model performance was increased with the addition of hC or H. Trait composition demonstrated distinct patterns between associated model configurations, where species that prefer intermediate to slow-flowing current conditions in regions further downstream performed better with bC–H. Including hydrological variables in SDMs contributes to improved performance, it is however, species-specific and future studies would benefit from hydrology-related variables to link environmental conditions and diverse communities. Consequently, SDMs that include climatic and hydrological variables could more accurately guide sustainable river ecosystem management.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Species distribution models (SDMs) are ecological predictive models, increasingly used to inform and complement large-scale distribution analyses to aid conservation efforts (Araújo et al., 2011; Guisan et al., 2013; Eaton et al., 2018). In river ecosystems, SDMs have been less often applied due, in part, to (1) complex interactions between the numerous driving factors of river systems and (2) insufficient abiotic and biotic data to effectively describe hydro-ecological relationships in streams.
The hydrological regime is said to be the “master variable” (Power et al., 1995) of lotic habitats and critical to the ecological stability of river ecosystems (Poff et al., 1997). It is highly variable, both spatially and temporally, making it a core driver of the physical structure of river habitats and a regulator of species distribution and abundance (Resh et al., 1988; Poff et al., 1997). For many river species, flow regimes are very important for, at least, part of their life history (Lytle & Poff, 2004). Numerous macroinvertebrate species depend on flow-related cues that directly or indirectly initiate, for example, breeding period (Hancock & Bunn, 1997), development (Gray, 1981) and emergence & metamorphosis (Peckarsky et al., 2000; Lytle, 2002). Therefore, species have evolved to the heterogeneity in rivers caused by the variability in flow regime, e.g. average/low/high flows, intermittent and ephemeral flows. It is essential to understand the influence stream flow has on the distribution of species, so that suitable recommendations can be made to restore or conserve river systems successfully. This is particularly important with the increasing changes in hydrological regime due to climate change, e.g. severity and frequency of floods and droughts (IPCC, 2007; Döll & Zhang, 2010; Kiesel et al., 2019; Gudmundsson et al., 2021).
Recently, data describing stream ecosystems in greater detail are becoming available, e.g. stream specific climate and land use (Domisch et al., 2015a; Linke et al., 2019), hydrology (Barbarossa et al. 2018; Irving et al. 2018), river classification and characteristics (Vogt et al., 2007; Ouellet Dallaire et al., 2019) as well as dams and reservoirs (globaldamwatch.org; Lehner et al., 2011). Despite this increasing availability, most studies focus on climate (e.g. Domisch et al., 2011; Ihlow et al., 2012; Markovic et al., 2014; Ruiz-Navarro et al., 2016; Kärcher et al., 2019; Rodríguez-Merino et al., 2019) and/or implement precipitation variables as surrogates of hydrological variables (e.g. Maloney et al., 2013; Pletterbauer et al., 2015; Domisch et al., 2019). While climate is certainly a dominant factor in driving species distribution and abundance, its sole use may be misrepresenting the effect on species’ distributions due to correlating factors, such as topography, which may impact predictions, leading to ambiguous conclusions (Real et al., 2013).
There are still only a limited number of studies that directly investigate the influence of flow regime on riverine species distribution. Some recent attempts have been made to include, at least, some aspects of flow regime, e.g. high flow days (Kuemmerlen et al., 2015a) as well as aggregated flow statistics, e.g. mean annual flow (Kuemmerlen et al., 2015b; Pyne & Poff, 2017), which were shown to be of high relevance to macroinvertebrate distribution. With data describing climate and hydrology becoming increasingly available it becomes possible to include both aspects to SDMs. However, the question remains, if including both will affect model results, leading to improved predictive accuracy and/or differing predicted ranges? Considering both climate and hydrology variables in SDMs would possibly allow for a distinct examination of the abiotic drivers of species distribution and facilitate management decisions to develop wise conservation or restoration strategies.
We hypothesized that SDMs including hydrological variables next to commonly used climatic variables would improve SDM performance (H1). To investigate this, we applied SDMs on a community of benthic macroinvertebrates with combinations of three predictor datasets including (1) climate only variables, (2) hydrology only variables and (3) both climate and hydrology-related variables. We evaluated the influence of each predictor dataset on species' distribution by investigating individual variance as well as shared variance explained by each dataset. We also compared differences in the functional traits; current preference and stream zonation preference. By investigating trait composition of, for example, species’ that perform better with hydrology variables, we aimed at better interpreting the model’s results, from a perspective closely related to river hydrology. We investigated which model configuration performed better by evaluating differences in model performance. Further, we analysed how the choice of predictor datasets influences the predicted species distributions. By comparing differences in model performance, functional traits and explained variance, we expected to determine the individual influence of both climate and hydrology datasets, and to what extent these datasets influence predicted species' distributions. If combinations of datasets that included hydrology outperformed the climate only combinations, we would accept H1. If performance across datasets was similar, H1 would be rejected.
Materials and methods
Study area
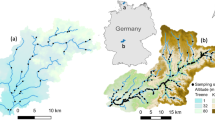
The study area comprised the Ems (17,934 km2) and Weser (46,306 km2) catchments located in Germany (Fig. 1). Selecting two catchments fully within Germany ensured consistent data availability. The two catchments are adjacent to each other and both are split into two distinct ecoregions: central plains (lowland) and central highlands (mountain, sensu Illies 1967).
The stream network for the study area was based on a layer in GeoTIFF raster format of a modeled 1 km2 gridded stream network with a total of 13,749 cells. The network was downloaded from earthenv.org/streams, which was derived from Hydrosheds (www.hydrosheds.org; 30-arc-s spatial grain; Lehner et al., 2008), which in turn is based on the SRTM dataset (srtm.csi.cgiar.org; Jarvis et al., 2008). This spatial resolution was applied due to the requirement of spatially analogous environmental variables in SDMs, i.e. hydrology, bioclimate and hydroclimate at 1 km2 (Araújo et al., 2019).
Biological data
Macroinvertebrate species datasets were obtained from German Federal State agencies. These macroinvertebrate samples were collected using a 25 × 25 cm hand net (500 μm mesh size), following the AQEM STAR sampling methodology, in which samples consist of 20 microhabitats, sampled based on their relative cover at the sampling site (AQEM 2002; Haase et al., 2004). To be included in the study, each species had to be identified to species level and to avoid issues with modelling a small sample size, each species had to have at least 20 occurrences within the study area (Stockwell & Townsend Peterson, 2002). A total of 91 species occurrences at 1,258 sites remained from this process, in a presence only format. Sampling frequency occurs in a 3-year cycle, all sites were sampled at least once within the period between 2005 and 2013.
Environmental predictors
The predictor variables for bioclimate, hydroclimate and hydrology used in this study were all openly sourced data and freely available to the user. All datasets are available in raster GeoTIFF format.
Bioclimate
There are 19 bioclimatic variables openly available from worldclim.org (Hijmans et al., 2005; Lehner et al., 2008; Fick & Hijmans, 2017) describing temperature and precipitation. These data are applied frequently in SDMs and other predictive modelling studies. The variables were downloaded at a 1 km (30-arc-s) spatial resolution. All 19 variables were masked to the base layer 1 km2 stream network described above. The bioclimate data were measured at local grid cell scale and represents our measurement of climate.
Hydroclimate
The hydroclimate variables were downloaded from earthenv.org/streams (Domisch et al., 2015a) and are based on, among others, the 19 bioclimatic variables described above. However, it differs in that the bioclimatic information is related to the stream network by accumulating information from the upper subcatchment in every point (i.e. grid-cell) along the stream network. Flow accumulation was used as the mechanism to relate these environmental variables with the stream network (see Domisch et al., 2015a for details). This accumulative feature causes high correlation among many of the variables and with stream flow (Kuemmerlen et al., 2014, 2015a) and hence includes an aspect of hydrological information. This dataset therefore contains information describing both climate and hydrology, embedded within the values.
Hydrology
The dataset from Irving et al. (2018), includes 53 of the Indicators of Hydrological Alteration (IHA) metrics that describe the magnitude, frequency, timing, duration and rate of change of flow events (Olden & Poff, 2003). IHA metrics are commonly used in flow-ecology assessments (e.g. Kakouei et al., 2018) and environmental flow research (Poff et al., 2010; Peres & Cancelliere, 2016) as they comprehensively describe hydrological flow regime. These metrics have only recently been included in river SDMs (but see Irving et al., 2020). As the information contained in the IHA metrics is directly related to the hydrological regime of rivers, it is logical to suggest that this data could affect predicted species’ distributions.
In brief, stream flow was extrapolated for the German stream network through a weighted linear regression using accumulated seasonal precipitation from earthenv.org/streams (Domisch et al., 2015a). The daily stream flow (m3 s−1) was then applied as input to calculate the IHA metrics via the R package Eflowstats (www.github.com/USGS-R/EflowStats; Henriksen et al., 2006; Archfield et al., 2014). These data were purposefully created at the same spatiotemporal resolution as the bioclimate and hydroclimate data for explicit use in SDMs. The data have been found to be effective for use in predictive modelling (Irving et al., 2018) within the same study area (Irving et al., 2020). All 53 IHA metrics were computed for the time period 1985–2013 to capture enough variability to produce accurate metrics following Kennard et al. (2010) who recommends at least 15 year’s worth of data, and informs that there is negligible change in variability over 30 years. In addition, we wanted to ensure the hydrological data covered both the biological (2005–2013) and bioclimate (1950–2000) data.
These metrics were originally derived from both gauging station daily stream flow data and seasonal precipitation sourced from earthenv.org/streams (Domisch et al., 2015a). It is therefore important to note that some degree of correlation is to be expected with the hydroclimate variables used in this study. Nonetheless, the additional high-resolution stream flow data adequately adds various aspects of flow regime at a high temporal resolution and therefore better represents hydrology.
Model set-up
Three predictor sets containing variables that describe (1) climate (bioclimate, bC; n = 19), (2) flow regime (hydrology, H; n = 53), (3) climate and hydrology combined (hydroclimate, hC; n = 19) were applied in this study. Hence, we set-up five model configurations to compare all eventualities: (1) bC only (bC; n = 19), (2) hydroclimate + hydrology (hC–H; n = 72), (3) bioclimate + hydrology (bC–H; n = 72), (4) hydroclimate + bioclimate (hC–bC; n = 38), and (5) hydroclimate + bioclimate + hydrology (hC–bC–H; full model: n = 91, descriptions in Table S1). By comparing the explained variance of each predictor set based on variance partitioning analysis, we identified the influence of each predictor set (bC, hC and H) on the spatial distribution of macroinvertebrates. Here, the full model (hC–bC–H) represents the full coverage of environmental predictors used in our study (n = 0) therefore, it is intended for comparative purposes only.
Variable selection process
The variables were selected by adapting the procedure from Irving et al. (2020) using boosted regression trees (BRTs), in a two-step process. First, each predictor set was applied in BRTs separately (hydroclimate n = 19, bioclimate n = 19, hydrology n = 53) for every species within the community (n = 91). BRTs calculate the variable importance of each predictor from the number of times each variable was chosen by the algorithm (Elith et al., 2008). The variable importance was averaged (mean) across all species to find the variable importance for the community. The average variable importance was used to determine the most important individual variables (i.e. 30% of variables with the highest relative importance) from each predictor set (hydroclimate n = 6, bioclimate n = 6, hydrology n = 19). The remaining variables from all predictor sets (n = 31) were then applied collectively into the 2nd run of BRTs with the same criteria as above.
A pair-wise Pearson’s correlation analysis was undertaken for each model configuration with the threshold 0.7 (Dormann et al., 2013). The variable importance from the 2nd run of BRTs was used to determine which of the correlated variables were chosen to remain in the analysis. In order to compare the datasets effectively, we retained a minimum of two predictor variables per dataset. Variables chosen for each model are outlined in Tables 1 and 2. As the variables are related, i.e. hydroclimate was derived from the bioclimate dataset, and hydrology was derived, in part, from the hydroclimate dataset, it was likely that a high level of correlation would be observed between datasets (see Table S2 for correlation matrix).
The selected variables (n = 9) were included in our final full model (hC–bC–H). The variables contained in the full model were then distributed according to the remaining model configurations: bC (n = 5), hC–H (n = 7), bC–H (n = 7), hC–bC (n = 7).
Species distribution models
All SDM analyses were undertaken in the R package sdm (Naimi & Araujo, 2016). We applied the four model configurations outlined above to each species within the community in separate SDMs. Each SDM was applied with an ensemble of five algorithms: artificial neural network (ANN), generalized linear model (GLM), flexible discriminant analysis (FDA), BRT, and classification tree analysis (TREE). As the species occurrence data consisted of presences only, we applied 2,000 randomly placed pseudo-absences in geographical space as background absences. Each model was repeated 10 times by bootstrapping. This resulted in 50 models per species, per configuration. For validation, the data were randomly split into training and testing datasets in 70:30 ratios. The true skills statistic (TSS) and the sensitivity values were derived from the validation as a measurement of model performance. Sensitivity is a measure of true positives, i.e. where the model correctly places a presence, and the TSS is derived from both the sensitivity and the specificity (the number of true negatives). The TSS values are reported as weighted mean ± standard error (mean ± SE). In addition, TSS values for individual species were used as a grouping measure for the functional traits analysis (see below).
To predict probability of occurrence for each species and model configuration, an ensemble model was produced by averaging all 50 previously mentioned individual models (5 algorithms × 10 repetitions) weighted by each model’s performance as given by the TSS values. This weighting emphasizes the higher performing models, without excluding lower performing models (Araújo & New, 2007). As output, the ensemble model produced a probability of occurrence map for the entire stream network of the study area. To convert the probability map to binary presence/absence predictions (1, 0), we applied a threshold determined from maximising sensitivity and specificity (Liu et al., 2005, 2013).
Statistical analysis
Model performance
All analyses were undertaken in R version 3.5.2 (R Core Team, 2018). Model performance was analyzed using the TSS values calculated through the training and testing validation datasets. Pairwise Wilcoxon tests were applied to the 50 TSS values of each species to test for differences between model configurations. Any values P < 0.05 were considered significantly different. Each configuration modelled 91 species resulting in a total of 364 Wilcoxon tests. We summarize the outcome as a percentage of significance for each model configuration, i.e. % S = (number of significant models/364) * 100.
Predicted distribution
The predicted distributions were compared according to range size and percentage overlap. Range size was defined as the number of raster cells predicted by the model as a presence within the study area, after converting the predicted probabilities to binary presence/absence predictions by maximizing the sensitivity and specificity produced by the ensemble prediction (Liu et al., 2005, 2013). Range size was determined by counting the number of species presences predicted by each model configuration. To test for differences in range size between model configurations, pairwise Wilcoxon tests were applied. To compare the predicted distribution of the community, and how they were similar or different in geographical space, pairwise range overlap values were calculated by counting the number of grid cells that contained a same species’ presence predicted by each respective model configuration. The proportion of shared grid cells in relation to the predicted range of the pairwise model configurations, was calculated as percentage overlap (mean ± SE) for the community. The predicted distributions of all species per model configuration were combined and projected into the study area in QGIS version 3.4 (QGIS.org, 2021) as a measure of species richness. Pairwise Wilcoxon tests were performed on the species richness of each model configuration.
Variance partitioning
Variance partitioning analysis was applied on all 2-set model configurations for each species. First, GLMs were performed on all binary predictions (i.e. presence/absence) to determine the proportional variance for each predictor set separately, then collectively according to the model configuration to ascertain the shared variance. Due to the nature of logistic regression, i.e. GLM, the standard coefficient of determination (R2) cannot be derived from the model. Therefore, from each GLM a pseudo R2 value was calculated through the Nagelkerke function in the rcompanion package (Mangiafico, 2019) using the McFadden method (de Araujo et al., 2014). It is important to note that pseudo R2 values cannot be interpreted in same manner as other regression techniques, i.e. ordinary least squares (OLS) where R2 can be interpreted as the amount of variance in the response variable explained by the predictor variable. The pseudo R2 value in GLM context is a relative measure between models of the same type describing how well the model explains the data (http://rcompanion.org/handbook/G_10.html, online book, accessed 25/09/2020). The pseudo R2 values were used here to determine the proportional contribution of variance as explained by the model, the total variance explained by the model being 1. This proportion was calculated following de Araujo et al. (2014), where the proportion of variance explained by the first predictor set can be described as total variance explained minus the proportion of variance explained by the second predictor set in the configuration.
The shared variance of both predictor sets can be described as the total variance explained minus the sum of both predictor sets in the configuration. For example, the proportional variance of the hydroclimate data in the hydroclimate and hydrology configuration would be; R2hydroclimate = 1 − R2hydrology, the proportional variance of hydrology; R2hydrology = 1 − R2hydroclimate, and the amount of shared variance: R2shared = 1 − (R2hydrology + R2hydroclimate). These proportional variance values were then used as input into the varPart function in modEva package (Barbosa et al., 2016) to calculate the proportional variance partition. This procedure was applied to every species and proportional variance was averaged across all species and reported as community proportional variance.
Analysis of traits
To better understand the influence of the different datasets on the composition of species’ communities, we grouped species according to their performance for each configuration based on TSS values from the SDMs. The highest TSS value determined a model configuration group for each species, that is, if 5 species showed higher TSS values for the bC configuration than all other configurations, those species were grouped as “bC”.
We compared the patterns of species response using functional traits describing; (1) stream zonation preference, and (2) current preference. These traits were chosen as they’ve been successfully used in a similar application and have high data availability (Irving et al., 2020). Trait information was downloaded from www.freshwaterecology.info (Bauernfeind et al., 1995; Eder et al., 1995; Graf et al., 1995a, 1995b, 2002a, 2002b, 2008, 2016–2019a, 2016–2019b; Hörner et al., 1995; Jäch et al., 1995; Janecek et al., 1995a, 1995b, 2002; Moog, 1995; Nesemann & Moog, 1995; Nesemann & Reischütz, 1995a, 1995b; Zettel, 1995; Schmedtje & Colling, 1996; AQEM, 2002; Bauernfeind et al., 2002; Buffagni et al., 2009; Schmidt-Kloiber & Hering, 2015; Schmidt-Kloiber et al., 2017; Buffagni et al., 2016–2019). Stream zonation are available in a 10-point assignment system in which each species was assigned 10 points distributed across the trait categories according to their preferences (e.g. rheophil, limno-rheophil). Current preference was available in binary format (1, 0) according to species preference. To standardize traits across models, trait values were converted to percentages as a measure of trait composition.
The percentage of trait composition for each group (e.g. bC group, bC–H group) were compared visually. In order to disentangle the influence of each dataset on species traits, we did not include the full model in this analysis. To compare trait composition to predictor variables, we applied pairwise Wilcoxon tests of difference to compare values located at the species occurrences associated with each group.
Results
Variable selection process
The variable selection process resulted in nine predictors: five bioclimate, two hydroclimate and two hydrology variables (Table S3 for BRT coefficients). These were applied in the full model configuration and distributed to each 2-set model (Table 1). Interestingly, variables describing mean temperature of both wettest quarter and of driest quarter from both the hydroclimate and the bioclimate predictor sets were included in the model (see Table 1). It could be expected that the same variable from both climate-related predictor sets would be highly correlated, however the correlation was negligible: Bioclimate/Hydroclimate 08 from bC and hC, corr = 0.37, Bioclimate/Hydroclimate 09 from bC and hC, corr = 0.30 (Table S2). Mean values for all chosen predictor variables are outlined in Table 2.
Model performance
The SDMs performed well over all (mean ± SE: bC; TSS = 0.58 ± 0.02, S = 0.79 ± 0.02, hC–H; TSS = 0.55 ± 0.02, S = 0.76 ± 0.02, bC–H; TSS = 0.61 ± 0.02, S = 0.80 ± 0.02, hC–bC; TSS = 0.60 ± 0.02, S = 0.79 ± 0.02, hC–bC–H; TSS = 0.62 ± 0.02, S = 0.80 ± 0.02, Table S4; Fig. 2). The percentage of significance for model configuration hC–bC–H (36%, n = 142) was greater than all remaining model configurations, followed by (in order of decreasing performance); bC–H (26.4%, n = 98), hC–bC (20.3%, n = 81), bC (12.9%, n = 40) then hC–H (6.3%, n = 16). See Table 3 for pairwise totals of significantly better models/species and % of significance.
Comparison of TSS and sensitivity values across five models (variable combinations): bC; bioclimate only, hC–H; hydroclimate and hydrology, bC–H; bioclimate and hydrology, hC–bC; hydroclimate and bioclimate, hC–bC–H; hydroclimate, bioclimate and hydrology. Boxplots (bar = median, box = IQR, whiskers = 1.5 × IQR and outliers)
Variance partitioning
We applied variance partitioning to the predicted distributions from the SDMs (presence/absence). The variance partitioning of the 2-set model configurations showed that bioclimate also had the highest explained variance compared with hydroclimate (0.4, Fig. 3a) and hydrology (0.57, Fig. 3b). Hydrology showed the lowest amount of explained variance in all 2-set models, compared with hydroclimate (0.16, Fig. 3c) and bioclimate (0.09, Fig. 3b). Hydroclimate showed lower explained variance than bioclimate (0.12, Fig. 3a) but higher explained variance than hydrology (0.44, Fig. 3c). The shared variance in all 2-set models was relatively low (Fig. 3a–c), the highest being between Bioclimate and Hydroclimate (0.1, Fig. 3a). This demonstrates that the predictor sets have a separate influence on species’ distribution.
Model configurations hC–H and bC–H showed a negative shared variance meaning that the two predictor sets explain the variance in different directions, i.e. both a positive and negative relationship. Unexplained variance was lowest in bC–H model configuration (0.36, Fig. 3b), compared with hC–H (0.44, Fig. 3a) and hC–bC (0.38, Fig. 3c).
Predicted distributions
Model configurations predicted similar range sizes (mean no. of presences; bC–H; 1,949.8 ± 159, bC; 1,917.1 ± 150.6, hC–H; 1,939.5 ± 131.1, hC–bC; 1,827.4 ± 139.5, hC–bC–H; 1,916 ± 145.9, Fig. 4). Pairwise Wilcoxon tests showed a significant difference (P = 0.02), between predicted range sizes of hC–H vs. hC–bC.
Distribution of all species predicted by each model: a bC; bioclimate only, b hC–H; hydroclimate and hydrology, c bC–H; bioclimate and hydrology, d hC–bC; hydroclimate and bioclimate, and e hC–bC–H; hydroclimate, bioclimate and hydrology. Points represent locations, colours represent number of species predicted presence at point locations
However, differences were apparent in the geographical location of predicted ranges as shown by the range overlap. The smallest percentage range overlap was between model configurations bC and hC–H (42.3 ± 1.6%, n = 945, Table 4). The largest range overlap was between model configurations bC and hC–bC (73.2 ± 1.3%, n = 1,477, Table 4). The differences in predicted ranges are shown when mapping species richness (Fig. 4) in geographical space. The pairwise Wilcoxon tests on species richness showed a significant difference between hC–H and all other model configurations (P < 0.001). In addition a significant difference was found between bC and hC–bC–H (P = 0.03) and a near significant difference between bC and bC–H (P = 0.08).
Analysis of traits
We compared the influence of model configurations groups; bC, bC–H and hC–bC on species trait composition. Group hC–H was removed from this analysis as it comprised only three species, hence was not data sufficient to investigate patterns in trait composition. There then remained models configuration groups of 10 species in bC, 44 species in bC–H and 37 species for hC–bC. This analysis derived some noteworthy patterns (Fig. 5). Trait composition for current preference showed rheophil and rheo-limophil (Fig. 5a) comprised species from model group bC (both 30% trait composition) and hC–bC (both 27% trait composition). Species in model group bC–H showed a high trait composition within limo-rheophil category (36%). Trait composition for stream zonation (Fig. 5b) for all model configurations tended to increase from upstream to downstream until a peak was reached, then the values decreased further downstream. Distinct peaks were associated with some models, i.e. bC (20%) and hC–bC (18%) species peaked within lower-trout region, whereas bC–H (20%) peaked in barbel region further downstream. Patterns of trait composition from current preference and stream zonation are comparable in that bC and hC–bC species were associated with higher current velocity and regions further upstream in comparison to bC–H species that were associated with lower current velocity and downstream regions. Trait composition for all models showed a high association for littoral region, i.e. bC (17%), bC–H (26%) and hC–bC (16%). The Wilcoxon tests of difference between species occurrence associated with each model configuration group showed a significant difference between groups bC and bC–H for predictors; Bioclimate 04 (mean ± SE; bC = 628 ± 0.4, bC–H = 619.6 ± 0.3, P = 0.01) and Bioclimate 08 (mean ± SE; bC = 12.3 ± 0.1, bC–H = 14.5 ± 0.05, P = 0.0005).
Discussion
We compared three datasets, combined in five dataset configurations, to evaluate their influence on macroinvertebrate distribution predictions and traits using SDMs. We found that the dataset bC had the most influence on the model in terms of proportional variance, however model performance was increased with the addition of hC or H. After the full model, the configuration bC–H performed best, while hC–H configuration performed least well. The different model configurations predicted similar range sizes, however were not always consistent in geographical space. The composition of species’ functional traits, demonstrated distinct patterns between the species with and without the dataset describing hydrology.
Variable selection
One precipitation variable was included in each model configuration, i.e. precipitation seasonality (Bioclimate 15, Tables 1, 2) from the bioclimate dataset. The inclusion of this variable indicates that precipitation-related data are important to include in SDMs as a separate entity to hydrology-related parameters (such as high flow volume), suggesting that local precipitation should not be used as a substitute for hydrology-related data (e.g. Domisch et al., 2019). This is supported by the high variance explained individually, as well as the low amount of shared explained variance demonstrated in all model configurations through variance partitioning.
Model performance and variance partitioning
The full model performed best overall. The full model, however, was only used a comparison in our study, therefore from the model configurations containing 1 or 2 datasets the model bC–H performed best. We therefore argue that these two predictor sets (bC and H) complement each other well and that adding hydrology into the model configuration can improve model performance, while keeping the number of predictor variables low. The only model configuration that did not contain bC variables, i.e. hC–H, performed least well overall, confirming the importance of including climate when evaluating species distribution.
Variance partitioning of the individual predictor sets both suggest that bC, i.e. spatially static temperature and precipitation-related variables, have the most influence on the studied macroinvertebrate community, at this scale. Nonetheless, hC and H, i.e. stream and flow-related variables improved model performance, even though they demonstrated a smaller influence in terms of variance partitioning. Our finding is not consistent with current flow-ecology theory that suggests hydrology (H) to be as important as climate (bC) in determining species distribution (Pyne & Poff, 2017).
The scale (13,749 km2) and resolution (1 km2 grid cells) at which the predictors are applied may partially explain these results (Pearson & Dawson, 2003; Randin et al., 2009; Lenoir et al., 2013; Domisch et al., 2015b; Record et al., 2018) as it may be too coarse to fully capture the influence of all the dimensions of the hydrological regime on macroinvertebrate distribution. The IHA metric MH21; high flow volume is the discharge of the highest flow, represents the more extreme aspects of hydrological regime. High flow volume is likely related to catchment size, i.e. peak flows driven by climate and geology, at large-scales (i.e. catchment scale) (Poff, 1997). At small scales, i.e. reach scale, large-scale variables describing the hydrological regime, such as flood and droughts, are able to induce changes in river biota communities by influencing small-scale habitat, e.g. hydraulics, riverbed substrates and stream channel morphology (Allen & Vaughn, 2010; Soranno et al., 2010). However, local-scale hydraulics, e.g. pool/riffles, and their resultant impact on physical microhabitats, e.g. creation of refugia, influence the distribution of biota, which reduces the influence of large-scale drivers (Poff, 1997; Frissell et al., 1986). Of the few freshwater SDM studies that include variables directly describing hydrology, together with climate, most show a high importance of either both climate and hydrology, or hydrology as the most importance variable (Bond et al., 2011; Kuemmerlen et al., 2014; Huang et al., 2016), although a lower influence of hydrology has been documented in stream headwaters (Monbertrand et al., 2019). Hydrology is certainly important at finer spatial scales (Kuemmerlen et al., 2014), and climate becomes more important with increasing scale (Friedrichs-Manthey et al., 2020). This scale-dependency is a recognised challenge in SDM research (Domisch et al., 2015b; Friedrichs-Manthey et al., 2020). Nonetheless, hydrology metric RA3; Fall Rate, describing the rate of negative changes in flow between consecutive days, can drive potential drought stress. This negatively affects different communities in the ecosystem such as local plant communities (The Nature Conservancy, 2009), and initiates cascading effects like resource scarcity. Therefore, even on large-scales, factors impacting local-scale changes may still be influential.
In addition, the disproportionate number of variables could drive the differences in explained variance, that is, five variables were applied from the Bioclimate dataset, whereas two variables were applied from each of Hydroclimate and Hydrology datasets. Although we ensured at least two variables from each dataset for a robust comparison, the remaining variable selection was chosen by the selection process. As the selection process had a high influence in determining the most important variables for this community and study area, we believe this approach is less biased than forcing the exact same number of variables from each dataset.
Despite these challenges, incorporating hydrology at the scale of this study does not hinder predictive ability (Araújo et al., 2019) and resulted in an improvement in SDM performance compared with climate alone. Applying the same predictors at a smaller spatial (e.g. reach scale) with finer resolution (e.g. < 100 m2: Kuemmerlen et al., 2014) may result in hydrology demonstrating a stronger influence (Friedrichs-Manthey et al., 2020), on macroinvertebrate distribution, however further investigation would be needed for confirmation.
Predicted distributions
The model configuration bC–H predicted the largest range size overall. Conversely, hC–bC predicted the smallest range size. However, we only found a significant difference in range size between model configurations hC–H (the model that performed least well) and hC–bC.
Despite similar range size, some model configurations predicted species’ distributions in different geographical locations as suggested by the range overlap and differences in species richness between model configurations. The ranges predicted by model configuration hC–H overlap between 42 and 57% with all other models and were significantly different to all other configurations in terms of species richness. The correlation analysis and variance partitioning suggests that the datasets contain distinct environmental information, rendering the differences in predicted range size and location plausible. Here, the SDMs are relating different environmental conditions to the species’ known occurrences and predicting suitable habitat accordingly. Nonetheless, as hC–H performed least well, and does not include the most important dataset (bC) it is reasonable to suggest that these predicted ranges do not contain enough relevant information for accurate predictions.
For other model configurations 67–77% range of each model overlapped. We found a significant difference in species richness between our most basic model (bC) and the full model (hC–bC–H). This difference may be driven by the addition of hydrology-related variables (i.e. hC and H datasets), or it may be driven by the full model containing the most information, hence higher complexity (Araújo et al., 2019). As the full model, in this study, was evaluated for comparison purposes only, it is challenging to disentangle the reason behind this result. Considering the near significance of the differences between bC and bC–H it is possible that the addition of pure hydrology variables (H) influences species’ predicted range, however we were unable to confirm this conclusion with our analysis.
Analysis of traits
The models that contained specific information on hydrology (bC–H) performed best when applied on species that prefer intermediate to slow-flowing current conditions in regions that were further downstream. Species modelled with more information regarding climate (bC, hC–bC) showed preference for high to intermediate flow conditions located further upstream. Model configurations hC–bC and bC–H were comparable in terms of model performance, explained variance and predicted ranges. However, the species that performed better with each configuration, demonstrate distinct differences in their trait composition. These findings agree with Irving et al. (2020) where species that were modelled with additional descriptive hydrology variables prefer relatively slower current conditions and downstream regions.
Variables describing climate (mean temperature of wettest quarter and temperature seasonality) varied significantly for species occurrences grouped in bC and bC–H models, with higher temperature and lower seasonality shown for bC–H. These differences suggest that the variability in climate is strong enough upstream to drive the distribution of certain macroinvertebrate species, however in downstream regions, hydrology plays a much larger role. It is possible that the species located in upstream regions, at least in relative terms, are better adapted to colder temperatures than species located downstream (Monbertrand et al., 2019), and hence may respond more strongly to climate than hydrology.
Flow regime in both the Ems and Weser catchments is generally driven by precipitation (i.e. pluvial) with seasonal high flows driven by snowmelt and precipitation (Koeniger et al., 2009). In the Weser catchment, increasing low flows as well as decreasing low flow duration have been documented over the last few decades (Bormann and Pinter, 2017). Although increased precipitation due to climate change may have influence (Hisdal et al., 2001), increased flows are likely attributed to reservoir management (Bormann & Pinter, 2017). The climate datasets (hC and bC) applied in this study may therefore be unable to capture changes in flow due to anthrogenic influences. In contrast, the hydrology dataset is derived, among other variables, from discharge gauging stations, which measure real-time stream flow (m3 s−1), and therefore account, to some extent, hydrological variability not driven by precipitation, e.g. anthropogenic alteration, urban run-off and groundwater influence. The hydrologic variables (and the anthropogenic disturbance to hydrological regimes) are likely to assist in better depicting the varied influences on the physical habitat of the river biota (Resh et al., 1988; Poff et al., 1997), which shapes the structure and function of river ecosystems. In contrast, the climate datasets (bC and hC) are applied as indirect surrogates for water temperature (Moore et al., 2005; Caissie, 2006) and hydrology (e.g. Domisch et al., 2019).
It is therefore plausible that the species with upstream preferences, may be located in areas with limited influence of reservoir management (i.e. upstream of reservoirs), and hence performed better with climate-related variables. In contrast, species downstream of reservoirs are more influenced by direct measures of hydrology. However, more investigation is needed to confirm this theory.
Role of hydrology in SDMs: implication for practice
Our findings have important implications when applying such models to inform conservation efforts: omitting flow regime variables in SDMs may lead to an underrepresentation of macroinvertebrate species that are sensitive to changes in flow. For example, SDMs applied under current, and future climate conditions have predicted range shifts of e.g. macroinvertebrate distribution to higher altitudes (Domisch et al. 2011, 2013). Adding complementary factors describing flow regime may result in more accurate predicted range, both for current and future potential distributions, subsequently requiring adjustments in mitigation strategies for conservation efforts. Many rivers in Germany (and globally) are highly altered from their natural regime. Water as a resource is in high demand continuously abstracted, recycled and discharged back into the system, or rivers serve as heavily managed waterways. By focusing on climate or climate-related surrogates, predicting species ranges may not capture the full diversity of functional traits, and hence may ignore important relationships between flow-sensitive species and their environment. This factor is especially important when predicting species’ distributions under future conditions, as it can provide insights to the ultimate drivers of range shifts (Monbertrand et al., 2019; Irving et al., 2020), and also should be considered when applying SDMs on other aquatic species (Ihlow et al., 2011; Markovic et al., 2014; Ruiz-Navarro et al., 2016; Rodriguez-Merino et al., 2019).
Concluding remarks
The main findings of our study suggest that by including environmental predictors describing flow regime in SDMs applied on macroinvertebrates can potentially increase model performance, despite a low contribution of hydrology to explained variance. The IHA metrics applied in this study are partially derived from real-time stream flow data from gauging stations, which incorporate the principal factors that control river hydrology. The metrics describe direct influencing factors of river habitat, including some aspects of anthropogenic disturbance. These characteristics are not described by either of the climate-related predictors included in our study. In addition, our analysis of functional traits provides insights into the driving factors of macroinvertebrate distribution. Improvement in model performance is species-specific and likely linked to the relevance of hydrology to particular species.
Our study intentionally focused on climatic and hydrologic variables however, we acknowledge that to provide a holistic assessment of benthic macroinvertebrate distribution, additional elements known to influence river systems need to be considered. It is therefore possible that the inclusion of data describing the surrounding land use (that influences stream flow and nutrient influx), topography (that influences river hydraulics) and connectivity (that influences dispersal and habitat availability) of the study area could increase predictive accuracy and influence range size.
Our study highlights improvements to predictive ability, and provides insights into drivers of community composition through functional traits. We recommend that given its fundamental importance, variables describing flow regime must be considered in SDM studies applied on river biota.
Data availability
Environmental datasets are available open access. Biological data were collected from German Federal State agencies and not publicly available.
Code availability
Not applicable.
References
Allen, D. C. & C. C. Vaughn, 2010. Complex hydraulic and substrate variables limit freshwater mussel species richness and abundance. Journal of the North American Benthological Society 29: 383–394.
AQEM, 2002. Ecological Classifications. AQEM Expert Consortium [available on internet at www.aqem.de].
Araújo, M. B. & M. New, 2007. Ensemble forecasting of species distributions. Trends in Ecology and Evolution 22: 42–47.
Araújo, M. B., D. Alagador, M. Cabeza, D. Nogués-Bravo & W. Thuiller, 2011. Climate change threatens European conservation areas. Ecology Letters 14: 484–492.
Araújo, M. B., R. P. Anderson, A. Márcia Barbosa, C. M. Beale, C. F. Dormann, R. Early, R. A. Garcia, A. Guisan, L. Maiorano, B. Naimi, R. B. O’Hara, N. E. Zimmermann & C. Rahbek, 2019. Standards for distribution models in biodiversity assessments. Science Advances 5: eaat4858.
Archfield, S. A., J. G. Kennen, D. M. Carlisle & D. M. Wolock, 2014. An objective and parsimonious approach for classifying natural flow regimes at a continental scale. River Research and Applications 30: 1166–1183.
Barbarossa, V., M. A. J. Huijbregts, A. H. W. Beusen, H. E. Beck, H. King & A. M. Schipper, 2018. FLO1K, global maps of mean, maximum and minimum annual streamflow at 1 km resolution from 1960 through 2015. Scientific Data 5: 180052.
Barbosa, A. M., J. A. Brown, A. Jimenez-Valverde & R. Real, 2016. modEvA: model evaluation and analysis. R package version 1.3.2 [available on internet at https://CRAN.R-project.org/package=modEvA]. Accessed 20 Mar 2019.
Bauernfeind, E., O. Moog & P. Weichselbaumer, 1995. Ephemeroptera. In Moog, O. (ed.), Fauna Aquatica Austriaca Lieferung 1995 Wasserwirtschaftskataster Bundesministerium für land- und Forstwirtschaft. Wien [Aquatic Fauna of Austria; Edition 1995–2002; Water Management Cadastre]. Federal Ministry of Agriculture, Forestry, Environment and Water Management, Vienna.
Bauernfeind, E., O. Moog & P. Weichselbaumer, 2002. Ephemeroptera. In Moog, O. (ed.), Fauna Aquatica Austriaca Lieferung 2002 Wasserwirtschaftskataster Bundesministerium für land- und Forstwirtschaft Umwelt und Wasserwirtschaft. Wien [Aquatic Fauna of Austria; Edition 1995–2002; Water Management Cadastre]. Federal Ministry of Agriculture, Forestry, Environment and Water Management, Vienna.
Bond, N., J. Thomson, P. Reich & J. Stein, 2011. Using species distribution models to infer potential climate change-induced range shifts of freshwater fish in south-eastern Australia. Marine and Freshwater Research 62: 1043–1061.
Bormann, H. & N. Pinter, 2017. Trends in low flows of German rivers since 1950: comparability of different low-flow indicators and their spatial patterns. River Research and Applications 33: 1191–1204.
Buffagni, A., D. Armanini, M. Cazzola, J. Alba-Tercedor, M. López-Rodríguez, J. Murphy, L. Sandin & A. Schmidt-Kloiber, 2016–2019. Dataset Ephemeroptera [available on internet at www.freshwaterecology.info – the taxa and autecology database for freshwater organisms]. Accessed 12 September 2021.
Buffagni, A., M. Cazzola, M. López-Rodríguez, J. Alba-Tercedor & D. Armanini, 2009. Distribution and Ecological Preferences of European Freshwater Organisms – Ephemeroptera, Vol. 3. Pensoft Publishers, Sofia-Moscow
Caissie, D., 2006. The thermal regime of rivers: a review. Freshwater Biology 51: 1389–1406.
de Araujo, C. B., L. O. Marcondes-Machado & G. C. Costa, 2014. The importance of biotic interactions in species distribution models: a test of the Eltonian noise hypothesis using parrots. Journal of Biogeography 41: 513–523.
Döll, P. & J. Zhang, 2010. Impact of climate change on freshwater ecosystems: a global-scale analysis of ecologically relevant river flow alterations. Hydrology and Earth System Sciences 14: 783–799.
Domisch, S., S. C. Jähnig & P. Haase, 2011. Climate-change winners and losers: stream macroinvertebrates of a submontane region in Central Europe. Freshwater Biology 56: 2009–2020.
Domisch, S., M. B. Araújo, N. Bonada, S. U. Pauls, S. C. Jähnig & P. Haase, 2013. Modelling distribution in European stream macroinvertebrates under future climates. Global Change Biology 19: 752–762.
Domisch, S., G. Amatulli & W. Jetz, 2015a. Near-global freshwater-specific environmental variables for biodiversity analyses in 1 km resolution. Scientific Data 2: 150073.
Domisch, S., S. C. Jahnig, J. P. Simaika, M. Kuemmerlen & S. Stoll, 2015b. Application of species distribution models in stream ecosystems: the challenges of spatial and temporal scale, environmental predictors and species occurrence data. Fundamental and Applied Limnology 186: 45–61.
Domisch, S., M. Friedrichs, T. Hein, F. Borgwardt, A. Wetzig, S. C. Jähnig & S. D. Langhans, 2019. Spatially explicit species distribution models: a missed opportunity in conservation planning? Diversity and Distributions 25: 758–769.
Dormann, C. F., J. Elith, S. Bacher, C. Buchmann, G. Carl, G. Carre, J. R. G. Marquez, B. Gruber, B. Lafourcade, P. J. Leitao, T. Munkemuller, C. McClean, P. E. Osborne, B. Reineking, B. Schroder, A. K. Skidmore, D. Zurell & S. Lautenbach, 2013. Collinearity: a review of methods to deal with it and a simulation study evaluating their performance. Ecography 36: 27–46.
Eaton, S., C. Ellis, D. Genney, R. Thompson, R. Yahr & D. T. Haydon, 2018. Adding small species to the big picture: species distribution modelling in an age of landscape scale conservation. Biological Conservation 217: 251–258.
Eder E., W. Hödl, O. Moog, H. Nesemann & M. K. W Pöckl, 1995. Crustacea. In Moog, O. (ed.), Fauna Aquatica Austriaca Lieferungen 1995–2002 Wasserwirtschaftskataster Bundesministerium für land- und Forstwirtschaft Umwelt und Wasserwirtschaft. Wien [Aquatic Fauna of Austria; Edition 1995–2002; Water Management Cadastre]. Federal Ministry of Agriculture, Forestry, Environment and Water Management, Vienna.
Elith, J., J. R. Leathwick & T. Hastie, 2008. A working guide to boosted regression trees. Journal of Animal Ecology 77: 802–813.
Fick, S. E. & R. J. Hijmans, 2017. WorldClim 2: new 1-km spatial resolution climate surfaces for global land areas. International Journal of Climatology 37: 4302–4315.
Friedrichs-Manthey, M., S. D. Langhans, T. Hein, F. Borgwardt, H. Kling, S. C. Jähnig & S. Domisch, 2020. From topography to hydrology – the modifiable area unit problem impacts freshwater species distribution models. Ecology and Evolution 10: 2956–2968.
Frissell, C. A., W. J. Liss, C. E. Warren & D. Hurley, 1986. A hierarchical framework for stream habitat classification: viewing streams in a watershed context. Environmental Management 10: 199–214.
Graf, W., U. Grasser & J. Waringer, 1995a. Trichoptera. In Moog, O. (ed.), Fauna Aquatica Austriaca Lieferung 1995 Wasserwirtschaftskataster Bundesministerium für land- und Forstwirtschaft. Wien [Aquatic Fauna of Austria; Edition 1995–2002; Water Management Cadastre]. Federal Ministry of Agriculture, Forestry, Environment and Water Management, Vienna.
Graf, W., U. Grasser & A. Weinzierl, 1995b. Plecoptera. In Moog, O. (ed.), Fauna Aquatica Austriaca Lieferung 1995 Wasserwirtschaftskataster Bundesministerium für land- und Forstwirtschaft Wien [Aquatic Fauna of Austria; Edition 1995–2002; Water Management Cadastre]. Federal Ministry of Agriculture, Forestry, Environment and Water Management, Vienna.
Graf, W., U. Grasser & J. Waringer, 2002a. Trichoptera. In Moog, O. (ed.), Fauna Aquatica Austriaca Lieferung 2002 Wasserwirtschaftskataster Bundesministerium für Bundesministerium für land- und Forstwirtschaft Umwelt und Wasserwirtschaft. Wien [Aquatic Fauna of Austria; Edition 1995–2002; Water Management Cadastre]. Federal Ministry of Agriculture, Forestry, Environment and Water Management, Vienna.
Graf, W., U. Grasser & A. Weinzierl, 2002b. Plecoptera. In Moog, O. (ed.), Fauna Aquatica Austriaca Lieferung 2002 Wasserwirtschaftskataster Bundesministerium für land- und Forstwirtschaft Umwelt und Wasserwirtschaft Wien [Aquatic Fauna of Austria; Edition 1995–2002; Water Management Cadastre]. Federal Ministry of Agriculture, Forestry, Environment and Water Management, Vienna.
Graf, W., J. Murphy, J. Dahl, C. Zamora-Muñoz & M. López-Rodríguez, 2008. Distribution and Ecological Preferences of European Freshwater Organisms – Trichoptera, Vol. 1. Pensoft Publishers, Sofia:
Graf, W., A. Lorenz, J. T. de Figueroa, S. Lücke, M. López-Rodríguez, J. Murphy & A. Schmidt-Kloiber, 2016–2019a. Dataset Plecoptera [available on internet at www.freshwaterecology.info – the taxa and aut- ecology database for freshwater organisms]. Accessed 12 September 2021.
Graf, W., J. Murphy, J. Dahl, C. Zamora-Muñoz, M. López-Rodríguez & A. Schmidt-Kloiber, 2016–2019b. Dataset Trichoptera [available on internet at www.freshwaterecology.info – the taxa and autecology database]. Accessed 12 September 2021.
Gray, L. J., 1981. Species composition and life histories of aquatic insects in a lowland Sonoran Desert stream. The American Midland Naturalist 106: 229–242.
Gudmundsson, L., J. Boulange, H. X. Do, S. N. Gosling, M. G. Grillakis, A. G. Koutroulis, M. Leonard, J. Liu, H. Müller Schmied, L. Papadimitriou, Y. Pokhrel, S. I. Seneviratne, Y. Satoh, W. Thiery, S. Westra, X. Zhang & F. Zhao, 2021. Globally observed trends in mean and extreme river flow attributed to climate change. Science 371: 1159–1162.
Guisan, A., R. Tingley, J. B. Baumgartner, I. Naujokaitis-Lewis, P. R. Sutcliffe, A. I. T. Tulloch, T. J. Regan, L. Brotons, E. McDonald-Madden, C. Mantyka-Pringle, T. G. Martin, J. R. Rhodes, R. Maggini, S. A. Setterfield, J. Elith, M. W. Schwartz, B. A. Wintle, O. Broennimann, M. Austin, S. Ferrier, M. R. Kearney, H. P. Possingham & Y. M. Buckley, 2013. Predicting species distributions for conservation decisions. Ecology Letters 16: 1424–1435.
Haase, P., S. Lohse, S. Pauls, K. Schindehütte, A. Sundermann, P. Rolauffs & D. Hering, 2004. Assessing streams in Germany with benthic invertebrates: development of a practical standardised protocol for macroinvertebrate sampling and sorting. Limnologica 34: 349–365.
Hancock, M. A. & S. E. Bunn, 1997. Population dynamics and life history of Paratya australiensis Kemp, 1917 (Decapoda: Atyidae) in upland rainforest streams, south-eastern Queensland, Australia. Marine and Freshwater Research 48: 361–369.
Henriksen, J. A., J. Heasley, J. G. Kennen & S. Nieswand, 2006. Users' manual for the Hydroecological Integrity Assessment Process software (including the New Jersey Assessment Tools). viii: 72.
Hijmans, R. J., S. E. Cameron, J. L. Parra, P. G. Jones & A. Jarvis, 2005. Very high resolution interpolated climate surfaces for global land areas. International Journal of Climatology 25: 1965–1978.
Hisdal, H., K. Stahl, L. M. Tallaksen & S. Demuth, 2001. Have streamflow droughts in Europe become more severe or frequent? International Journal of Climatology 21: 317–333.
Hörner, K., O. Moog & F. Sporka, 1995. Oligochaeta. In Moog, O. (ed.), Fauna Aquatica Austriaca Lieferungen 1995, 2002 Wasserwirtschaftskataster Bundesministerium für land- und Forstwirtschaft Umwelt und Wasserwirtschaft Wein [Aquatic Fauna of Austria; Edition 1995, 2002; Water Management Cadastre]. Federal Ministry of Agriculture, Forestry, Environment and Water Management, Vienna.
Ihlow, F., J. Dambach, J. O. Engler, M. Flecks, T. Hartmann, S. Nekum, H. Rajaei & D. Rödder, 2012. On the brink of extinction? How climate change may affect global chelonian species richness and distribution. Global Change Biology 18: 1520–1530.
Illies, J., 1967. Limnofauna Europaea: Stuttgart 1967; Prospekt, Gustav Fisher Verlag, Stuttgart.
IPCC, 2007. In Solomon, S., D. Qin, M. Manning, Z. Chen, M. Marquis, K. B. Averyt, M. Tignor & H. L. Miller (eds), Climate Change 2007: The Physical Science Basis. Contribution of Working Group I to the Fourth Assessment Report of the Intergovernmental Panel on Climate Change. Cambridge University Press, Cambridge.
Irving, K., M. Kuemmerlen, J. Kiesel, K. Kakouei, S. Domisch & S. C. Jähnig, 2018. A high-resolution streamflow and hydrological metrics dataset for ecological modeling using a regression model. Scientific Data 5: 180224.
Irving, K., S. C. Jähnig & M. Kuemmerlen, 2020. Identifying and applying an optimum set of environmental variables in species distribution models. Inland Waters 10: 11–28.
Jäch, M., J. Kodada, O. Moog, W. Paill & S. Schödl, 1995. Coleoptera. In Moog, O. (ed.), Fauna Aquatica Austriaca Lieferungen 1995–2002 Wasserwirtschaftskataster Bundesministerium für land- und Forstwirtschaft Umwelt und Wasserwirtschaft Wien [Aquatic Fauna of Austria; Edition 1995–2002; Water Management Cadastre]. Federal Ministry of Agriculture, Forestry, Environment and Water Management, Vienna.
Janecek, B., O. Moog, C. Moritz, C. Orendt & R. Saxl, 1995a. Chironomidae. In Moog, O. (ed.), Fauna Aquatica Austriaca Lieferung 1995 Wasserwirtschaftskataster Bundesministerium für land- und Forstwirtschaft. Wien [Aquatic Fauna of Austria; Edition 1995, 2002; Water Management Cadastre]. Federal Ministry of Agriculture, Forestry, Environment and Water Management, Vienna.
Janecek, B., O. Moog & J. Waringer, 1995b. Odonata. In Moog, O. (ed.), Fauna Aquatica Austriaca Lieferungen 1995, 2002 Wasserwirtschaftskataster Bundesministerium für land- und Forstwirtschaft Umwelt und Wasserwirtschaft. Wien [Aquatic Fauna of Austria; Edition 1995–2002; Water Management Cadastre]. Federal Ministry of Agriculture, Forestry, Environment and Water Management, Vienna.
Janecek, B., O. Moog, C. Moritz, C. Orendt & R. Saxl, 2002. Chironomidae. In Moog, O. (ed.), Fauna Aquatica Austriaca Lieferung 2002 Wasserwirtschaftskataster Bundesministerium für land- und Forstwirtschaft Umwelt und Wasserwirtschaft. Wien [Aquatic Fauna of Austria; Edition 1995, 2002; Water Management Cadastre]. Federal Ministry of Agriculture, Forestry, Environment and Water Management, Vienna.
Jarvis, A., H. I. Reuter, A. Nelson & E. Guevara, 2008. Hole-filled SRTM for the globe Version 4, available from the CGIAR-CSI SRTM 90m Database.
Kakouei, K., J. Kiesel, S. Domisch, K. S. Irving, S. C. Jähnig & J. Kail, 2018. Projected effects of climate-change-induced flow alterations on stream macroinvertebrate abundances. Ecology and Evolution 8: 3393–3409.
Kärcher, O., K. Frank, A. Walz & D. Markovic, 2019. Scale effects on the performance of niche-based models of freshwater fish distributions. Ecological Modelling 405: 33–42.
Kiesel, J., A. Gericke, H. Rathjens, A. Wetzig, K. Kakouei, S. C. Jähnig & N. Fohrer, 2019. Climate change impacts on ecologically relevant hydrological indicators in three catchments in three European ecoregions. Ecological Engineering 127: 404–416.
Kuemmerlen, M., B. Schmalz, B. Guse, Q. Cai, N. Fohrer & S. C. Jähnig, 2014. Integrating catchment properties in small scale species distribution models of stream macroinvertebrates. Ecological Modelling 277: 77–86.
Kuemmerlen, M., B. Schmalz, Q. Cai, P. Haase, N. Fohrer & S. C. Jähnig, 2015a. An attack on two fronts: predicting how changes in land use and climate affect the distribution of stream macroinvertebrates. Freshwater Biology 60: 1443–1458.
Kuemmerlen, M., S. Stoll, A. Sundermann & P. Haase, 2015b. Long-term monitoring data meet freshwater species distribution models: lessons from an LTER-site. Ecological Indicators 65: 122–132.
Lehner, B., K. Verdin & A. Jarvis, 2008. New global hydrography derived from spaceborne elevation data. EOS Transactions American Geophysical Union 89: 93–94.
Lehner, B., C. R. Liermann, C. Revenga, C. Vörösmarty, B. Fekete, P. Crouzet, P. Döll, M. Endejan, K. Frenken, J. Magome, C. Nilsson, J. C. Robertson, R. Rödel, N. Sindorf & D. Wisser, 2011. High-resolution mapping of the world’s reservoirs and dams for sustainable river-flow management. Frontiers in Ecology and the Environment 9: 494–502.
Lenoir, J., B. J. Graae, P. A. Aarrestad, I. G. Alsos, W. S. Armbruster, G. Austrheim, C. Bergendorff, H. J. B. Birks, K. A. Bråthen, J. Brunet, H. H. Bruun, C. J. Dahlberg, G. Decocq, M. Diekmann, M. Dynesius, R. Ejrnæs, J.-A. Grytnes, K. Hylander, K. Klanderud, M. Luoto, A. Milbau, M. Moora, B. Nygaard, A. Odland, V. T. Ravolainen, S. Reinhardt, S. M. Sandvik, F. H. Schei, J. D. M. Speed, L. U. Tveraabak, V. Vandvik, L. G. Velle, R. Virtanen, M. Zobel & J.-C. Svenning, 2013. Local temperatures inferred from plant communities suggest strong spatial buffering of climate warming across Northern Europe. Global Change Biology 19: 1470–1481.
Linke, S., B. Lehner, C. Ouellet Dallaire, J. Ariwi, G. Grill, M. Anand, G. Günther, P. Beames, V. Burchard-Levine, S. Maxwell, H. Moidu, F. Tan & M. Thieme, 2019. Global hydro-environmental sub-basin and river reach characteristics at high spatial resolution. Scientific Data 6: 283.
Liu, C., P. M. Berry, T. P. Dawson & R. G. Pearson, 2005. Selecting thresholds of occurrence in the prediction of species distributions. Ecography 28: 385–393.
Liu, C., M. White & G. Newell, 2013. Selecting thresholds for the prediction of species occurrence with presence-only data. Journal of Biogeography 40: 778–789.
Lytle, D. A., 2002. Flash floods and aquatic insect life-history evolution: evaluation of multiple models. Ecology 83: 370–385.
Lytle, D. A. & N. L. Poff, 2004. Adaptation to natural flow regimes. Trends in Ecology and Evolution 19: 94–100.
Maloney, K. O., D. E. Weller, D. E. Michaelson & P. J. Ciccotto, 2013. Species distribution models of freshwater stream fishes in Maryland and their implications for management. Environmental Modeling and Assessment 18: 1–12.
Mangiafico, S., 2019. rcompanion: Functions to Support Extension Education Program Evaluation. R package version 2.1.1 [available on internet at https://CRAN.R-project.org/package=rcompanion].
Markovic, D., S. Carrizo, J. Freyhof, N. Cid, S. Lengyel, M. Scholz, H. Kasperdius & W. Darwall, 2014. Europe’s freshwater biodiversity under climate change: distribution shifts and conservation needs. Diversity and Distributions 20: 1097–1107.
Monbertrand, A. L. B., P. Timoner, K. Rahman, P. Burlando, S. Fatichi, Y. Gonseth, F. Moser, E. Castella & A. Lehmann, 2019. Assessing the vulnerability of aquatic macroinvertebrates to climate warming in a mountainous watershed: supplementing presence-only data with species traits. Water (Switzerland) 11: 1–29.
Moog, O., 1995. Fauna Aquatica Austria – a comprehensive species inventory of Austrian aquatic organisms with ecological notes. In Forestry FMfAa. Wasserwirtschaftskataster, Vienna.
Moore, R. D., P. Sutherland, T. Gomi & A. Dhakal, 2005. Thermal regime of a headwater stream within a clear-cut, coastal British Columbia, Canada. Hydrological Processes 19: 2591–2608.
Naimi, B. & M. Araujo, 2016. SDM: a reproducible and extensible R platform for species distribution modelling. Ecography 39: 368–375.
Nesemann, H. & O. Moog, 1995. Hirudinea. In Moog, O. (ed.), Fauna Aquatica. Austriaca Lieferungen 1995–2002 Wasserwirtschaftskataster Bundesministerium für land- und Forstwirtschaft Umwelt und Wasserwirtschaft. Wein [Aquatic Fauna of Austria; Edition 1995–2002; Water Management Cadastre]. Federal Ministry of Agriculture, Forestry, Environment and Water Management, Vienna.
Nesemann, H. & P. Reischütz, 1995a. Bivalvia. In Moog, O. (ed.), Fauna Aquatica Austriaca Lieferungen 1995, 2002 Wasserwirtschaftskataster Bundesministerium für land- und Forstwirtschaft Umwelt und Wasserwirtschaft Wien [Aquatic Fauna of Austria; Edition 1995–2002; Water Management Cadastre]. Federal Ministry of Agriculture, Forestry, Environment and Water Management, Vienna.
Nesemann, H. & P. Reischütz, 1995b. Gastropoda. In Moog, O. (ed.), Fauna Aquatica Austriaca Lieferungen 1995, 2002 Wasserwirtschaftskataster Bundesministerium für land- und Forstwirtschaft Umwelt und Wasserwirtschaf. Wien.
Olden, J. D. & N. L. Poff, 2003. Redundancy and the choice of hydrologic indices for characterizing streamflow regimes. River Research and Applications 19: 101–121.
Ouellet Dallaire, C., B. Lehner, R. Sayre & M. Thieme, 2019. A multidisciplinary framework to derive global river reach classifications at high spatial resolution. Environmental Research Letters 14: 024003.
Pearson, R. G. & T. P. Dawson, 2003. Predicting the impacts of climate change on the distribution of species: are bioclimate envelope models useful? Global Ecology and Biogeography 12: 361–371.
Peckarsky, B. L., B. W. Taylor & C. C. Caudill, 2000. Hydrologic and behavioral constraints on oviposition of stream insects: implications for adult dispersal. Oecologia 125: 186–200.
Peres, D. J. & A. Cancelliere, 2016. Environmental flow assessment based on different metrics of hydrological alteration. Water Resources Management 30: 5799–5817.
Pletterbauer, F., A. H. Melcher, T. Ferreira & S. Schmutz, 2015. Impact of climate change on the structure of fish assemblages in European rivers. Hydrobiologia 744: 235–254.
Poff, N. L., 1997. Landscape filters and species traits: towards mechanistic understanding and prediction in stream ecology. Journal of the North American Benthological Society 16: 391–409.
Poff, N. L., J. Allan, M. Bain, J. Karr, K. Prestegaard, B. Richter, R. Sparks & J. Stromberg, 1997. The natural flow regime. Bioscience 47: 769–784.
Poff, N. L., B. D. Richter, A. H. Arthington, S. E. Bunn, R. J. Naiman, E. Kendy, M. Acreman, C. Apse, B. P. Bledsoe, M. C. Freeman, J. Henriksen, R. B. Jacobson, J. G. Kennen, D. M. Merritt, J. H. O’Keeffe, J. D. Olden, K. Rogers, R. E. Tharme & A. Warner, 2010. The ecological limits of hydrologic alteration (ELOHA): a new framework for developing regional environmental flow standards. Freshwater Biology 55: 147–170.
Power, M. E., A. Sun, G. Parker, W. E. Dietrich & J. T. Wootton, 1995. Hydraulic food-chain models. Bioscience 45: 159–167.
Pyne, M. I. & N. L. Poff, 2017. Vulnerability of stream community composition and function to projected thermal warming and hydrologic change across ecoregions in the western United States. Global Change Biology 23: 77–93.
QGIS.org, 2021. QGIS Geographic Information System. QGIS Association [available on internet at http://www.qgis.org].
R Core Team, 2018. R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing [available on internet at https://www.R-project.org/].
Randin, C. F., R. Engler, S. Normand, M. Zappa, N. Zimmermann, P. B. Pearman, P. Vittoz, W. Thuiller & A. Guisan, 2009. Climate change and plant distribution: local models predict high-elevation persistence. Global Change Biology 15: 1557–1569.
Real, R., D. Romero, J. Olivero, A. Estrada & A. L. Márquez, 2013. Estimating how inflated or obscured effects of climate affect forecasted species distribution. PLoS ONE 8: e53646.
Record, S., A. Strecker, M.-N. Tuanmu, L. Beaudrot, P. Zarnetske, J. Belmaker & B. Gerstner, 2018. Does scale matter? A systematic review of incorporating biological realism when predicting changes in species distributions. PLoS ONE 13: e0194650.
Resh, V. H., A. V. Brown, A. P. Covich, M. E. Gurtz, H. W. Li, G. W. Minshall, S. R. Reice, A. L. Sheldon, J. B. Wallace & R. C. Wissmar, 1988. The role of disturbance in stream ecology. Journal of the North American Benthological Society 7: 433–455.
Rodríguez-Merino, A., R. Fernández-Zamudio & P. García-Murillo, 2019. Identifying areas of aquatic plant richness in a Mediterranean hotspot to improve the conservation of freshwater ecosystems. Aquatic Conservation: Marine and Freshwater Ecosystems 29: 589–602.
Ruiz-Navarro, A., P. K. Gillingham & J. R. Britton, 2016. Predicting shifts in the climate space of freshwater fishes in Great Britain due to climate change. Biological Conservation 203: 33–42.
Schmedtje, U. & M. Colling. 1996. Ökologische typisierung der aquatischen makrofauna [Ecological Typing of Aquatic Macrofauna], Vol. 4. Informationsberichte des Bayerischen Landesamtes für Wasserwirtschaft.
Schmidt-Kloiber, A. & D. Hering, 2015. www.freshwaterecology.info – an online tool that unifies, standardises and codifies more than 20,000 European freshwater organisms and their ecological preferences. Ecological Indicators 53: 271–282.
Schmidt-Kloiber, A., P. Neu, M. Malicky, F. Pletterbauer, H. Malicky & W. Graf, 2017. Aquatic biodiversity in Europe: a unique dataset on the distribution of Trichoptera species with important implications for conservation. Hydrobiologia 797: 11–27.
Soranno, P. A., K. S. Cheruvelil, K. E. Webster, M. T. Bremigan, T. Wagner & C. A. Stow, 2010. Using landscape limnology to classify freshwater ecosystems for multi-ecosystem management and conservation. Bioscience 60: 440–454.
Stockwell, D. R. B. & A. Townsend Peterson, 2002. Effects of sample size on accuracy of species distribution models. Ecological Modelling 148: 1–13.
The Nature Conservancy, 2009. Indicators of Hydrologic Alteration Version 7.1 User's Manual.
Vogt, J., P. Soille, A. de Jager, E. Rimavičiūtė, W. Mehl, S. Foisneau, K. Bódis, J. Dusart, M. Paracchini, P. Haastrup & C. Bamps, 2007. A Pan-European River and Catchment Database, Luxembourg (Luxembourg).
Acknowledgements
This work has been supported by the German Federal Ministry of Education and Research (BMBF; 01LN1320A and 033W034A). We thank the German Federal State environmental agencies for providing biological data. For help with data collection and formatting we thank Melissa Schulte and Judith Mahnkopf. We appreciate the constructive comments by three reviewers that greatly improved the manuscript.
Funding
Open Access funding enabled and organized by Projekt DEAL. This work has been supported by the German Federal Ministry of Education and Research (BMBF; 01LN1320A and 033W034A).
Author information
Authors and Affiliations
Contributions
All authors contributed to the study conception and design. Material preparation, data collection and analysis were performed by KI. The first draft of the manuscript was written by KI and all authors commented on previous versions of the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
All authors declare that they have no conflict of interest or financial ties to disclose.
Additional information
Handling Editor: Verónica Ferreira
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Irving, K., Jähnig, S.C. & Kuemmerlen, M. Disentangling the effect of climatic and hydrological predictor variables on benthic macroinvertebrate distributions from predictive models. Hydrobiologia 849, 1021–1040 (2022). https://doi.org/10.1007/s10750-021-04765-w
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10750-021-04765-w