Abstract
Potato onion (Allium cepa var. aggregatum G. Don) remained the most widely grown onion type in Finland up to the 1950’s, after which the more productive cultivars of common onion replaced this vegetatively propagated onion type. The Finnish accessions have been maintained in two national plant genetic resources (PGR) collections since 1990’s but new samples have been obtained still later. In this research, we analysed the genetic diversity of potato onions and shallots, grown in home gardens and in PGR collection, and compared the uniformity of the two PGR collections. One hundred ten onion samples were analysed using eight microsatellite loci, which amplified 83 alleles in total. There was a large number of genetically different samples, forming four major groups in the dendrogram. The results show that there is still a lot of genetic diversity among the Finnish potato onions, even though they are propagated vegetatively. The rich diversity in the samples obtained from citizens reflects a valuable source of genetic resources maintained by home gardeners. The national collection has been updated on the basis of the results. To ensure the preservation of the rich onion heritage, efforts are needed to increase the availability of potato onion to home gardeners and commercial growers.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Allium cepa L. is a widely grown, economically important species which is propagated by seeds or bulbs. There is great variability within the species, e.g. in bulb characteristics, flavour, storage ability and in the ability to bolt or produce daughter bulbs in the first season. A. cepa is commonly classified to two groups: Common onion group, reproduced from seeds or from seed-grown sets, and to Aggregatum group which reproduces mainly via vegetative daughter bulbs (Fritsch and Friesen 2002).
Aggregatum group produces smaller bulbs than common onions and usually an aggregated cluster of bulbs. According to Fritsch and Friesen (2002), Aggregatum group comprises both shallots and potato onions which are difficult to distinguish from each other and intermediate forms exist. Compared to shallots, potato onions have typically larger bulb size, fewer and somewhat flattened daughter bulbs which remain enclosed by the skin of the mother bulb longer than in the shallots (Fritsch and Friesen 2002). However, Leino et al. (2018) reported that neither morphological nor genetic results support a division between potato onions and shallots. Generally, the variability within Aggregatum group is poorly represented in genebanks, because vegetatively propagated onions often carry latent viruses (Fritsch and Friesen 2002).
In northern Europe, potato onion is a traditional onion type, having a long cultivation history within families. It has also been called multiplying onion or shallot in some countries (Leino et al. 2018). In Finland, it has been documented that the most common cultivated onion in the early 1900’s was potato onion, called also “Russian onion” (Lundén 1921; Smirnoff 1904; Sunila et al. 1928), which refers to the origin of the onions. Potato onion was also commonly called “set onion” referring to its propagation way from bulbs (e.g. Lundén 1921). Potato onion remained the most widely grown onion type in Finland up to the 1950’s (Osara and Bremer 1989), after which the more productive cultivars of common onion replaced the vegetatively propagated potato onion. Aura (1963) collected and compared the morphological characteristics of 27 Finnish potato onion accessions. According to his analyses, the accessions were grouped into the more uniform North Finnish type, producing larger number of bulbs with smaller size, and into the South Finnish type, producing lower number of bulbs with bigger size. Aura (1963) suggested that most of the Finnish potato onions originate from onions imported from Russia before the World War I and the local accessions were developed from original seed-propagated material as the result of selection by the growers. However, some of the South Finnish accessions might have been imported from Central Europe, too.
In 1980’s, the interest in this traditional Finnish onion rose again, with the scope to compare the characteristics of different accessions and to eliminate viruses. Osara and Bremer (1989) collected 112 accessions, most of which originated from North Finland. This material forms the basis for the current Finnish PGR collection of potato onions. The collection has been maintained in two locations at Luke since 1990’s, one in southwestern Finland in Piikkiö and one in northern Finland (in Rovaniemi until 2015 and in Sotkamo since 2016). In 2011–2013, a new inventory of potato onions maintained in situ was performed (Heinonen 2014) which resulted in 45 contacts of potato onion growers. Some samples have been obtained from home gardeners even later.
Based on morphological traits, Osara and Bremer (1989) stated that the northern accessions were more uniform than accessions collected from southern or eastern Finland. However, there are difficulties in performing the characterisation of accessions using only morphological characters. Leino et al. (2018) reported large variation in different characters within the accessions and between different experiments, due to environmental factors, properties of the set onions and possible genotype-environment interaction. The only character showing a consistent scoring among experiments was the bulb skin colour (Leino et al. 2018).
Due to difficulties in identifying accessions using only morphological characters, new molecular methods are increasingly applied to study genetic diversity. There are several DNA markers of choice for this, e.g. microsatellite (SSR = simple sequence repeat) markers that have been developed (Baldwin et al. 2012; Fischer and Bachmann 2000; Jakše et al. 2005; McCallum et al. 2008) and used for estimating genetic diversity in onion. However, only two studies have been carried out in potato onion (Leino et al. 2018; Rungis et al. 2020).
The aim of this research was to analyse the genetic diversity of the Finnish potato onions, grown in home gardens and in the national PGR collection. We also aimed to compare the uniformity of the two collections, maintained in southern and northern Finland. The results were used for reforming the national PGR collection of potato onion to conserve high diversity and remove duplicate accessions.
Materials and methods
Plant material
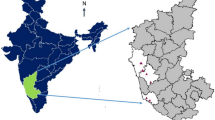
Samples which originated from the Finnish national collection of potato onion as well as from home gardens were analysed in two phases. In 2012, 35 samples were taken from the collection maintained at Luke’s experimental station in Piikkiö (60°25′30′′ N, 022°31′00′′ E) and in 2016, 29 samples from Luke’s duplicate collection in Sotkamo experimental station (64°07′50″ N, 28°23′0″ E). Principally, the same accessions were grown in both experimental stations and the aim was to check the genetic identity of these collections. In addition, 35 samples obtained from Finnish home gardens were included in the 2012 analysis. Sampling was based on a call which was made in 2012 and seeked for potato onions still maintained by home gardeners for several decades of known cultivation history (Heinonen 2014). After 2012, some new notices on old potato onions were still received from citizens and nine new samples were included in the 2016 analysis. The collection sites of the samples are shown in Fig. 1. One of the samples originated from Estonia. Samples, their collecting sites and analysis years are listed in Supplementary Information. Accessions which were included in the earlier genetic studies (Leino et al. 2018; Rungis et al. 2020) are also marked in Supplementary information.
Collection sites of onion samples obtained from citizens or the Finnish onion PGR collection. Size of the symbol refers to the number of samples from the same municipality. Letters refer to genetic groups in the dendrogram (Fig. 2)
Altogether, there were 110 different onion samples (Table 1), 64 from Luke’s national collection (28 accessions were analysed from both experimental stations), 44 from home gardens and 2 reference samples (common onion and red onion) from a commercial onion grower. Six of the accessions from the national PGR collection were assumed shallots, according to the information obtained at receiving the samples in early 1990’s. The protocols and equipment for analysing samples in the two different years were somewhat different, and the exact protocols are given only from the most recent study, differences in 2012 are mentioned in parenthesis.
E.Z.N.A® SP Plant DNA kit (Omega Bio-tek, Norcross, GA, USA) was used for DNA extractions from frozen leaves (2012: CTAB—Cetyl trimethylammonium bromide—extraction). DNA concentrations were measured using a NanoDrop™ 1000 Spectrophotometer (Thermo Fisher Scientific Ltd, Vantaa, Finland).
Microsatellite analyses
Microsatellites selected for the study were developed for A. cepa cv. Kaba (Fischer and Bachmann 2000). The seven selected ones were AMS06, AMS12, AMS13, AMS16, AMS23, AMS25, and AMS30 (Table 2).
To separate and visualise amplified products, an ABI PRISM® 310 Genetic Analyzer (Thermo Fisher Scientific Ltd, Vantaa, Finland) was used (2012: MegaBACE 500 Sequencer), and the forward primer of each primer pair was labelled with a fluorescent dye, FAM™ (5-carboxyfluorescein), VIC® or TET™ (6-carboxytetrachlorofluorescein). Microsatellites were amplified in a PCR reaction of 10 µl, containing 0.05 U Biotools polymerase (Biotools B&M Labs, S.A., Madrid, Spain), 1 µl buffer supplied by the enzyme manufacturer containing 2 mM MgCl2, 200 µM each dNTP, 20 ng of DNA, and 500 nM each primer. The PCR program for microsatellites AMS06, AMS13 and AMS 25 was as described in Fischer and Bachmann (2000). For the rest, the PCR amplification conditions were as follows: 37 cycles of 30 s at 94 °C, 45 s at 50 °C (AMS30), 55 °C (AMS16, and AMS23) or 60 °C (AMS12), and 60 s at 72 °C in a BioRad C1000 thermal Cycler (Bio-Rad, Hercules, California, USA). The program contained an initial denaturation step of 5 min at 94 °C and was followed by a final extension step of 5 min at 72 °C. PCR products were amplified separately but some with different fluorescent label were joined for the ABI runs (AMS12 + AMS13 and AMS23 + AMS30), dilutions differed from 1/20 to 1/200. GeneMapper® software 5 was used for allele size estimation.
Data analyses
Polymorphic information content (PIC) for microsatellite loci was counted with the program Cervus version 3.0.7 (Kalinowski et al. 2007) according to the formula by Bostein et al. (1980). DARwin software version 6.0.014 (Dissimilarity Analysis and Representation for Windows, Perrier and Jacquemoud-Collet 2006) was used for counting a dissimilarity index between samples based on the Dice coefficient. To visualise dissimilarities, an unweighted neighbor-joining (NJ, Saitou and Nei 1987) tree was built. In addition, principal coordinates analysis (PCoA) based on genetic distances between samples was performed using the software GenAlEx 6.4 (Peakall and Smouse 2006, 2012). GenAlEx was also used for counting genetic diversity indices H0 (observed heterozygosity) and Shannon’s diversity index I (Shannon 1948), to compare the samples from citizens with those from the collection.
Results
Eight microsatellite loci (primers for AMS12 amplified two different loci) were used for studying the genetic diversity among the Finnish potato onions and shallots. All the SSRs contained dinucleotide repeats and amplified 83 alleles in total (Table 1), the number of alleles in a locus varying from 2 to 17 (mean 10.3). PIC values varied from 0.36 to 0.84 (mean 0.72).
Genetic distances between accessions were visualised with an NJ tree (Fig. 2) and PCoA (Fig. 3). In the dendrogram (Fig. 2), four main groups were formed (A = 32 samples, B = 47 samples, C = 13 samples, and D = 4 samples), and the groups were split into subgroups. Twenty-eight accessions were analysed from duplicate PGR collections (2012 samples from Piikkiö and 2016 from Sotkamo, Table 1), and some of them differed in their microsatellite results: 11 slightly (1–2 allelic differences) and 3 greatly (GS3, PII_Lappeenranta HY 88B, and Ylämaa HY 90B). In the PCoA (Fig. 3) the first two axes explained 46% of the variation among the 96 samples. Samples of group A2 were most clearly grouped together.
The dendrogram containing 87 potato onions, 7 assumed shallots (marked with an asterisk), common onion, and red onion. Samples were analysed either in year 2012 or 2016 or both. The samples that were analysed in both years and were proven to be duplicates do not have the year marked after their name; those that differed from each other greatly are indicated with arrows. Samples from the existing plant genetic resource collection are written in capital letters
Some grouping based on collection sites (Fig. 1) could be observed: landraces from northern Finland were mainly in subgroup A1, which, however, also contained samples from other parts of Finland. Eastern accessions were mainly in subgroup A2. The biggest group B was very heterogeneous containing accessions from all over Finland.
The dendrogram (Fig. 2) revealed three bigger clonal groups containing 11–16 identical or nearly identical (with a maximum of two allele differences) accessions of which at least one was from the existing collection. These could represent old and more widely spread potato onion accessions which still are under cultivation. Genetic diversity was a little higher in the samples from the collection compared to those from citizens (number of alleles 47 vs. 41, Shannon’s information indexI1.642 vs. 1.535, Ho 0.620 vs. 0.497). However, rich variability was also still left within in situ samples, as 14 accessions received from the citizens (n = 44) differed with at least in three alleles from the existing samples in the PGR collection.
The reference samples, red onion and common onion, did not group in the dendrogram near to any potato onions reflecting the fact that they contained some alleles that did not exist in potato onions. Red onion was in group B and common onion in group C. All the six samples that were recognized as shallots, were in group B, and three of them were genetically identical (GS2, Sante2, and GS5). One of the assumed shallots, Sante1 was genetically identical to six potato onion accessions.
Discussion
The genetic results showed that there is still a lot of genetic diversity among the Finnish potato onions, even though they are propagated vegetatively. Our results are slightly divergent to the findings of Leino et al. (2018) who reported that there is a high degree of clonality in the Fennoscandian multiplying onions. However, they also found that the Finnish accessions showed higher diversity and differed from Scandinavian accessions. The material in the study by Leino et al. (2018) included only 14 Finnish accessions, three of which were probably breeding material and other three regarded as shallot varieties. Thus, our study gives a more comprehensive view to the genetic diversity within the Finnish cultivated potato onions.
Samples in our study were collected from different areas in Finland, indicating that potato onions have been grown all over the country during the last decades. Although some grouping of accessions according to geographical locations could be observed in the dendrogram, it is evident that onions have moved with the home gardeners to new growing sites, which was also reported by Leino et al. (2018). The geographic location marked in the Fig. 1 was the collection site, i.e. the most recent growing location, which may be far from the traditional growing area of the accession. As an example, the “Sample 40 Somero” was cultivated in the Southwest Finland (Somero 60°37′45″ N, 23°30′50″ E), but it was originally obtained from the southwestern part of Lapland (Keminmaa 65°48′05″ N, 24°32′40″ E), where it has been cultivated for several decades. The home gardener specifically wanted a northern potato onion from the locality where she was born.
Most of the citizens, who replied to the onion inventory in 2011–2013 (Heinonen 2014), knew the history of their onions one or two generations backwards in their family or they had received onions from friends, neighbours or other sources within Finland. Based on the respondents’ memories, we can assume that many in situ samples of potato onion have over one hundred years of growing history, but they have been often moved to new growing sites. Therefore, we cannot anymore separate North and South Finnish potato onion types, as Aura (1963) did in his report in 1960’s.
In the European study by Rungis et al. (2020), focusing on the genetic diversity of European potato onion collection, most of the Finnish accessions were unique in comparison with other samples. Only two of the 24 Finnish samples represented genotypes that were also found in other Nordic countries. The Finnish samples in the study were selected to represent separate genotypes according to our preliminary data. Therefore, they included a large share of the genetic diversity available in the Finnish PGR collection at the time of the study. According to Rungis et al. (2020), the Finnish accessions were genetically intermediate between the Nordic and Baltic groups. The high genetic variation and uniqueness might be related to the suggested origin of the Finnish potato onions from seed-propagated material from Russia (Lundén 1921; Aura 1963). This cannot be, however, confirmed without further analyses, including material from larger geographical area.
One of the accessions in our study, Sante1, which was regarded as a shallot, was genetically identical to six potato onion accessions. However, the division between potato onions and shallots seems to be unclear (Fritsch and Friesen 2002), and according to Leino et al. (2018) it might be mainly related to linguistic traditions without any clear genetic distinction.
Twenty-eight samples in the study were from duplicate collections (Piikkiö and Sotkamo) and some of them gave different microsatellite results. Minor differences in microsatellites might be due to different methods and equipment used and to different interpretations (two different persons). On the other hand, major differences, observed in three accessions, are likely to be caused by sample mixing over the years, as the samples had been grown in different locations. This reflects the challenges in the practical maintenance of field collections over years and highlights the need to check the genetic identity of duplicate collections.
The rich diversity in the samples obtained from citizens reflects a valuable source of genetic resources maintained by home gardeners. Most of the variability was already present in the existing PGR collection but also new accessions differing greatly from the samples in the collection could be found. According to the present results the collection has been updated. Six new accessions were taken to the collection which now contains 29 accessions with the status accepted and a few with the pending status.
There is a rising interest in using old domestic crop plants both among home gardeners and small-scale commercial growers. However, potato onions are often heavily infected by viruses which hinders the use of accessions in commercial propagation and production. In 1980’s, Osara and Bremer (1989) and Bremer et al. (1991) found that all accessions under study carried viruses, the most common of which were onion yellow dwarf virus, OYDV, and shallot latent virus, SLV. In vitro cultivation methods can be used to produce healthy bulbs, but this is time-consuming and expensive. To ensure the preservation of the high genetic diversity and to enhance the utilisation of the accessions, more research efforts should be laid on virus elimination and safe cultivation procedures to ensure good plant health in the whole production chain. Moreover, efforts to increase the availability of potato onion to both home gardeners and commercial growers would ensure the preservation of our rich onion heritage.
Availability of data and materials
The datasets generated during the study are available from the corresponding author on reasonable request.
References
Aura K (1963) Studies on the vegetatively propagated onions cultivated in Finland, with special reference to flowering and storage. Ann Agr Fenn 2(suppl. 5):1–74
Baldwin S, Pither-Joyce M, Wright K, Chen L, McCallum J (2012) Development of robust genomic simple sequence repeat markers for estimation of genetic diversity within and among bulb onion (Allium cepa L.) populations. Mol Breed 30:1401–1411
Bostein D, White RL, Skolnick M, Davis RW (1980) Construction of a genetic linkage map in man using restriction fragment length polymorphisms. Am J Hum Genet 32:314–331
Bremer K, Osara K, Palohuhta JP (1991) Syötävien sipulikasvien tervetaimituotanto. Tutkimuksen loppuraportti. Report, University of Helsinki
Fischer D, Bachmann K (2000) Onion microsatellites for germplasm analysis and their use in assessing intra- and interspecific relatedness within the subgenus Rhizirideum. Theor Appl Genet 101:153–164
Fritsch RM, Friesen N (2002) Evolution, domestication and taxonomy. In: Rabinowitch HD, Currah L (eds) Allium crop science: recent advances. CAB International, Wallingford, pp 5–30
Heinonen M (2014) Landrace in situ conservation strategy for Finland. MTT Report 163. http://urn.fi/URN:ISBN:978-952-487-570-7
Jakše J, Martin W, McCallum J, Havey MJ (2005) Single nucleotide polymorphisms, indels, and simple sequence repeats for onion cultivar identification. J Am Soc Hort Sci 130:912–917
Kalinowski ST, Taper ML, Marshall TC (2007) Revising how the computer program CERVUS accommodates genotyping error increases success in paternity assignment. Mol Ecol 16:1099–1106
Leino MW, Solberg SØ, Tunset HM, Fogelholm J, Karlsson Strese E-M, Hagenblad J (2018) Patterns of exchange of multiplying onion (Allium cepa L. Aggregatum-Group) in Fennoscandian Home Gardens. Econ Bot 72:346–356. https://doi.org/10.1007/s12231-018-9426-2
Lundén O (1921) Keittiökasvikirja Suomen puutarhureita ja puutarhaopetusta varten. (Translated from Swedish to Finnish by Maiju Pöyhönen and Ilmari Ahma), 2nd edn. Kustannusyhtiö Otavan kirjapaino, Helsinki
McCallum J, Thomson S, Pither-Joyce M, Kenel F, Clarke A, Havey MJ (2008) Genetic diversity analysis and singlenucleotide polymorphism marker development in cultivated bulb onion based on expressed sequence tag—simple sequence repeat markers. J Am Soc Hort Sci 133:810–818
Osara K, Bremer K (1989) Ryvässipulin viljelyn elvytys ja viljelyn varmentaminen. Report, University of Helsinki
Perrier X, Jacquemoud-Collet JP (2006) DARwin software. http://darwin.cirad.fr/darwin
Peakall R, Smouse PE (2006) GENALEX 6: genetic analysis in Excel. population genetic software for teaching and research. Mol Ecol Notes 6:288–295
Peakall R, Smouse PE (2012) GenAlEx 6.5: genetic analysis in Excel. Population genetic software for teaching and research—an update. Bioinformatics 28:2537–2539
Rungis D, Leino MW, Lepse L, Goreta Ban S, de Vahl E, Annamaa K, Põldma P, Suojala-Ahlfors T, Juškevičienė D, Kik C, Vågen IM, Stavělíková H (2020) Genetic characterization of European potato onion (Allium cepa Aggregatum Group) collections. Genet Res Crop Evol. https://doi.org/10.1007/s10722-020-01014-2)
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425
Shannon CE (1948) A mathematical theory of communication. Bell Syst Tech J 27(379–423):623–656
Smirnoff A (1904) Käsikirja yksinkertaisessa puutarhanhoidossa kansaa varten. (Translated from Swedish to Finnish by Alli Nissinen). Helsingin sentraalikirjapaino, Helsinki
Sunila JE, Brander U, Nylander H (eds) (1928) Pienviljelijän käsikirja, 5th edn. Werner Söderström Osakeyhtiö, Porvoo, p 476
Acknowledgements
The authors wish to thank Marja-Riitta Arajärvi, Sirpa Moisander, Hannu Ojanen, Johanna Rihtilä, Jaana Sarlin and Anneli Virta for their excellent technical assistance. Finnish home gardeners are thanked for providing us the plant material.
Funding
Open access funding provided by Natural Resources Institute Finland (LUKE). Sampling of potato onions from home gardens and the SSR analysis in 2012 was implemented in PGR Secure project (Novel characterization of crop wild relative and landrace resources as a basis for improved crop breeding, EU 7th Framework Programme, GA No. 266394). SSR analysis in 2016 was funded by Finnish National Plant Genetic Resources Programme. Cultivation in situ details were complemented in Farmers’ pride project (Networking, partnerships and tools to enhance in situ conservation of European plant genetic resources, H2020 Framework Programme, GA No. 774271).
Author information
Authors and Affiliations
Contributions
All authors contributed to the design of the study. Material preparation, data collection and analyses were performed by Pirjo Tanhuanpää and Kristiina Antonius. The first draft of the manuscript was written by Terhi Suojala-Ahlfors, Maarit Heinonen and Pirjo Tanhuanpää. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Suojala-Ahlfors, T., Heinonen, M., Tanhuanpää, P. et al. Rich diversity in cultivated Finnish potato onions (Allium cepa var. aggregatum G. Don). Genet Resour Crop Evol 69, 1547–1555 (2022). https://doi.org/10.1007/s10722-021-01317-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10722-021-01317-y