Abstract
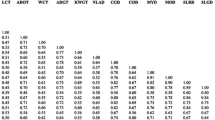
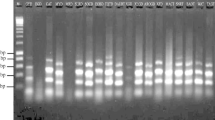
Coconut (Cocos nucifera L.) production sustains millions of smallholder farmers and their families around the globe, and also powers a thriving industry of climate-adaptive nutrition and biomaterials. Demand for high-quality planting materials has risen sharply in recent years, calling for greater understanding and potential utilization of the genetic diversity in coconut populations. Genetic diversity among several conserved coconut varieties in Vietnam was investigated to assess the suitability of 15 start codon targeted (SCoT) primers for future selective breeding programs. Amplification of 15 SCoT primers in 57 individuals from 19 coconut varieties revealed high intra-varietal diversity (Nei’s genetic diversity index = 0.237, Shannon’s Information index = 0.352), especially among the Vietnamese Dwarfs. Intriguingly, inter-varietal differentiation accounted for only 9.41% of total genetic variation. The designated SCoT primers were moderately informative. On average, each primer had 9.5 reproducible polymorphic bands per reaction. Polymorphism Information Content (PIC) per locus ranged between 0.217 and 0.390, with Resolving power (Rp) between 0.271 and 0.599. Principal Coordinate Analysis (PCoA) and Unweighted Pair Group Method with Arithmetic mean (UPGMA) analyses revealed distinct clusters for non-native Dwarf, native Dwarf, and Tall varieties. This paper represents the first-of-its-kind attempt to shed light on coconut genetic diversity in Vietnam using molecular markers.





Similar content being viewed by others
Data availability
Data supporting the findings of this study are available from the corresponding authors (Nguyen Bao Quoc and Nguyen Phuong Thao) upon reasonable request.
Abbreviations
- SCoT :
-
Start codon targeted
- QTL :
-
Quantitative trait locus
- PIC :
-
Polymorphism information content
- Rp :
-
Resolving power
- PCoA :
-
Principal coordinate analysis
- UPGMA :
-
Unweighted pair group method with arithmetic mean
References
Ashburner GR, Thompson WK, Halloran GM (1997) RAPD analysis of South Pacific coconut palm populations. Crop Sci 37(3):992–997
Baudouin L, Philippe R, Quaicoe R, Dery S, Dollet M (2009) General overview of genetic research and experimentation on coconut varieties tolerant/resistant to Lethal Yellowing. Oléagineux Corps Gras Lipides 16(2):127–131
Chempakam B, Ratnambal MJ (1993) Variation for leaf polyphenols in coconut cultivars. In Advances in Coconut Research and Development. Oxford and IBH Publishing Co. Pvt. Ltd., New Delhi, India, pp. 51–53.
Chen H, He XH, Luo C, Zhu J, Li F (2010) Analysis on the genetic diversity of 24 longan (Dimocarpus longan) accessions by SCoT markers. Acta Horticulturae Sinica 37:1651–1654
Collard BC, Mackill DJ (2009) Start codon targeted (SCoT) polymorphism: a simple, novel DNA marker technique for generating gene-targeted markers in plants. Plant Mol Biol Report 27(1):86
Dasanayaka PN, Nandadasa HG, Everard JMDT, Karunanayaka EH (2009) Analysis of coconut (Cocos nucifera L.) diversity using microsatellite markers with emphasis on management and utilisation of genetic resources. J Nat Sci Found Sri Lanka 37(2):99–109
De Riek J, Calsyn E, Everaert I, Van Bockstaele E, De Loose M (2001) AFLP based alternatives for the assessment of distinctness, uniformity and stability of sugar beet varieties. Theor Appl Genet 103(8):1254–1265
Duran Y, Rohde W, Kullaya A, Goikoetxea P, Ritter E (1997) Molecular analysis of East African tall coconut genotypes by DNA marker technology. J Genet Breed 51:279–288
Everard JMDT, Katz M, Gregg K (1996) Inheritance of RAPD markers in the coconut palm Cocos nucifera L. Trop Agric Res 8:124–138
Geethanjali S, Rukmani JA, Rajakumar D, Kadirvel P, Viswanathan PL (2018) Genetic diversity, population structure and association analysis in coconut (Cocos nucifera L.) germplasm using SSR markers. Plant Gen Res 16(2):156
Gunn BF, Baudouin L, Olsen KM (2011) Independent origins of cultivated coconut (Cocos nucifera L.) in the Old World tropics. PLOS ONE 6(6):21143
Guo X, Elston R (1999) Linkage information content of polymorphic genetic markers. Hum Hered 49(2):112–118
Hayati PK, Hartana A (2000) Genetic diversity of Jombang coconut population based on RAPD markers. In International Conference of Science and Technology for Managing Plant Genetic Diversity in the 21st Century, pp. 12–16.
Hennink S, Zeven AC (1990) The interpretation of Nei and Shannon-Weaver within population variation indices. Euphytica 51(3):235–240
Lebrun P, N’cho YP, Seguin M, Grivet L, Baudouin L (1998) Genetic diversity in coconut (Cocos nucifera L.) revealed by restriction fragment length polymorphism (RFLP) markers. Euphytica 101(1):103–108
Lebrun P, Baudouin L, Bourdeix R, Konan JL, Barker JH, Aldam C, Ritter E (2001) Construction of a linkage map of the Rennell Island Tall coconut type (Cocos nucifera L.) and QTL analysis for yield characters. Genome 44(6):962–970
Lewontin RC (1972) The apportionment of human diversity. Evol Biol 6:381–398
Loiola CM, Azevedo AON, Diniz LE, Aragao WM, Azevedo CDDO, Santos PHA, Ramos HCC, Pereira MG, Ramos SRR (2016) Genetic relationships among tall coconut palm (Cocos nucifera L.) accessions of the International Coconut Genebank for Latin America and the Caribbean (ICG-LAC), evaluated using microsatellite markers (SSRs). PloS one 11(3):0151309
Manimekalai R, Nagarajan P, Kumaran PM (2006) Comparison of effectiveness of RAPD, ISSR and SSR markers for analysis of coconut (Cocos nucifera L.) germplasm accessions. Trop Agric Res 18:217–226
Mauro-Herrera M, Meerow AW, Borrone JW, Kuhn DN, Schnell RJ (2007) Usefulness of WRKY gene-derived markers for assessing genetic population structure: an example with Florida coconut cultivars. Sci Hortic 115(1):19–26
Meerow AW, Wisser RJ, Brown SJ, Kuhn DN, Schnell RJ, Broschat TK (2003) Analysis of genetic diversity and population structure within Florida coconut (Cocos nucifera L.) germplasm using microsatellite DNA, with special emphasis on the Fiji Dwarf cultivar. Theor Appl Gen 106(4):715–726
Meerow AW, Noblick L, Borrone JW, Couvreur TL, Mauro-Herrera M, Hahn WJ, Kun DN, Nakamura K, Oleas NH, Schnell RJ (2009) Phylogenetic analysis of seven WRKY genes across the palm subtribe Attaleinae (Arecaceae) identifies Syagrus as sister group of the coconut. PLoS One 4(10):7353
Nei M (1978) Estimation of average heterozygosity and genetic distance from a small number of individuals. Genetics 89(3):583–590
Niral V, Jerard BA (2018) Botany, origin and genetic resources of coconut. In the coconut palm (Cocos nucifera L.)-research and development perspectives. Springer, Singapore
Oyoo ME, Muhammed N, Cyrus KN, Githiri SM (2016) Assessment of the genetic diversity of Kenyan coconut germplasm using simple sequence repeat (SSR) markers. Afr J Biotech 15(40):2215–2223
Peakall R, Smouse PE (2006) GENALEX 6: genetic analysis in excel. Population genetic software for teaching and research. Mol Ecol Notes 6:288–295
Peakall R, Smouse PE (2012) GENALEX 6.5: genetic analysis in Excel. Population genetic software for teaching and research-an update. Bioinformatics 28:2537–2539
Perera L, Russell JR, Provan J, McNicol JW, Powell W (1998) Evaluating genetic relationships between indigenous coconut (Cocos nucifera L.) accessions from Sri Lanka by means of AFLP profiling. Theor Appl Gen 96(3–4):545–550
Perera L, Russell JR, Provan J, Powell W (2000) Use of microsatellite DNA markers to investigate the level of genetic diversity and population genetic structure of coconut (Cocos nucifera L.). Genome 43(1):15–21
Perera L, Russell JR, Provan J, Powell W (2003) Studying genetic relationships among coconut varieties/populations using microsatellite markers. Euphytica 132(1):121–128
Preethi P, Rahman S, Naganeeswaran S, Sabana AA, Gangaraj KP, Jerard BA, Niral V, Rajesh MK (2020) Development of EST-SSR markers for genetic diversity analysis in coconut (Cocos nucifera L.). Mol Biol Rep 47(12):9385–9397
Prevost A, Wilkinson MJA (1999) New system of comparing PCR primers applied to ISSR fingerprinting of potato cultivars. Theor Appl Genet 98(1):107–112
Rajesh MK, Jerard BA, Preethi P, Thomas RJ, Karun A (2014) Application of RAPD markers in hybrid verification in coconut. Crop Breed Appl Biotech 14(1):36–41
Rajesh MK, Sabana AA, Rachana KE, Rahman S, Jerard BA, Karun A (2015) Genetic relationship and diversity among coconut (Cocos nucifera L.) accessions revealed through SCoT analysis. Biotech 5(6):999–1006
Rajesh MK, Sabana AA, Rachana KE, Rahman S, Ananda KS, Karun A (2016) Development of a SCoT-derived SCAR marker associated with tall-type palm trait in arecanut and its utilization in hybrid (dwarf x tall) authentication. Ind J Gen Plant Breed 76:119–122
Rajesh MK, Fayas TP, Naganeeswaran S, Rachana KE, Bhavyashree U, Sajini KK, Karun A (2016) De novo assembly and characterization of global transcriptome of coconut palm (Cocos nucifera L.) embryogenic calli using Illumina paired-end sequencing. Protoplasma 253(3):913–928
Rajesh MK, Karun A, Parthasarathy VA (2018) Coconut biotechnology. In the coconut palm (Cocos nucifera L.)-research and development perspectives. Springer, Singapore
Reynolds KB, Cullerne DP, El Tahchy A, Rolland V, Blanchard CL, Wood CC, Singh SP, Petrie JR (2019) Identification of genes involved in lipid biosynthesis through de novo transcriptome assembly from Cocos nucifera developing endosperm. Plant Cell Physiol 60(5):945–960
Rivera R, Edwards KJ, Barker JHA, Arnold GM, Ayad G, Hodgkin T, Karp A (1999) Isolation and characterization of polymorphic microsatellites in Cocos nucifera L. Genome 42:668–675
Rohde W, Kullaya A, Rodriguez MJ, Ritter E (1996) Genome analysis of Cocos nucifera L. by PCR amplification of spacer sequences separating a subset of Copia-like Ecorirepelitive elements. Philippine J Crop Sci (philippines) 21:26–31
Rohde W, Sniady V, Herran A, Estioko L, Sinje S, Marseillac N, Angelique B, Patricia L, Dieter P, Alois K, Judith R, Norbert B, Rodriguez J (2002) Construction and exploitation of high density DNA marker and physical maps in the perennial tropical oil crops coconut and oil palm: from biotechnology towards marker-assisted breeding. Burotrop Bulletin 20:13–14
Rohlf FJ (2002) NTSYSpc: Numerical taxonomy system, ver. 2.1. Setauket, NY: Exeter Publishing Ltd.
Saensuk C, Wanchana S, Choowongkomon K, Wongpornchai S, Kraithong T, Imsabai W, Chaichoompu E, Ruanjaichon V, Toojinda T, Vanavichit A, Arikit S (2016) De novo transcriptome assembly and identification of the gene conferring a “pandan-like” aroma in coconut (Cocos nucifera L.). Plant Sci 252:324–334
Shalini KV, Manjunatha S, Lebrun P, Berger A, Baudouin L, Pirany N, Raganath RM, Prasad DT (2007) Identification of molecular markers associated with mite resistance in coconut (Cocos nucifera L.). Genome 50(1):35–42
Spellerberg IF, Fedor PJA (2003) A tribute to Claude Shannon (1916–2001) and a plea for more rigorous use of species richness, species diversity and the ‘Shannon–Wiener’Index. Glob Ecol Biogeogr 12(3):177–179
Teulat B, Aldam C, Trehin R, Lebrun P, Barker JH, Arnold GM, Karp A, Baudouin L, Rognon F (2000) An analysis of genetic diversity in coconut (Cocos nucifera) populations from across the geographic range using sequence-tagged microsatellites (SSRs) and AFLPs. Theor Appl Genet 100(5):764–771
Upadhya A, Jayadev K, Manimekalai R, Parthasarathy VA (2004) Genetic relationship and diversity in Indian coconut accessions based on RAPD markers. Sci Hortic 99(3–4):353–362
Xiao Y, Luo Y, Yang Y, Fan H, Xia W, Annaliese S, Mason AS, Zhao S, Sager R, Qiao F (2013) Development of microsatellite markers in Cocos nucifera and their application in evaluating the level of genetic diversity of Cocos nucifera. Plant Omics 6(3):193–200
Yang Y, Bocs S, Fan H, Armero A, Baudouin L, Xu P, Xu J, This D, Hamelin C, Iqbal A, Qadri R, Zhou L, Li Z, Wu Y, Ma Z, Issali AE, Rivallan R, Liu N, Xia W, Peng M, Xiao Y (2021) Coconut genome assembly enables evolutionary analysis of palms and highlights signaling pathways involved in salt tolerance. Commun Biol 4(1):1–14
Yeh FC, Yang RC, Boyle TB, Ye ZH, Mao JX (1997) Popgene, version 1.32; the user-friendly shareware for population genetic analysis. Can Univ Alberta Mol Biol Biotechnol Centre 10:295–301
Zhang J, Guo D, Gong Y, Liu C, Li M, Zhang G (2011) Optimization of start codon targeted polymorphism PCR (SCoT-PCR) system in Vitis vinifera. J Fruit Sci 28(2):209–214
Zizumbo-Villarreal D, Ruiz-Rodriguez M, Harries H, Colunga-GarciaMarin P (2006) Population genetics, lethal yellowing disease, and relationships among mexican and imported coconut ecotypes. Crop Sci 46:2509–2516
Acknowledgements
We are grateful to the Dong Go Coconut Research Center for generous provision of leaf materials and information.
Funding
This research was funded by the Ministry of Science and Technology (MOST, Vietnam) under grant number ĐTĐL.CN-12/19.
Author information
Authors and Affiliations
Contributions
Conceptualization: NBQ, Data curation: NHMK, HNXM, NDNP, NBQ, Formal analysis: NHMK, Funding acquisition: NPT, Investigation: NHMK, NTQ, HNXM, NDNP, NPT and NBQ, Methodology: NBQ, NHMK, Validation: NBQ, NPT, Writing—original draft: NHMK, NBQ, Writing—review and editing: NBQ, NPT, NTQ.
Corresponding authors
Ethics declarations
Conflict of interest
The authors of this article have no conflicts of interest.
Consent for publication
All authors have given their consent for publication.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Khang, N.H.M., Quang, N.T., Mai, H.N.X. et al. Genetic characterization of coconut (Cocos nucifera L.) varieties conserved in Vietnam through SCoT marker-based polymorphisms. Genet Resour Crop Evol 69, 385–398 (2022). https://doi.org/10.1007/s10722-021-01237-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10722-021-01237-x