Abstract
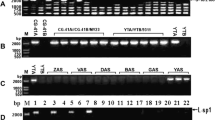
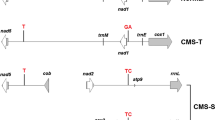
Cytoplasmic male sterility (CMS) has been exclusively used as an emasculation tool in seed production of hybrids in onion (Allium cepa L.).Three types of onion CMS (CMS-S, CMS-R, and CMS-T) have been used in hybrid onion breeding. Male sterility conferred by both CMS-S and CMS-R is likely induced by open reading frame (orf) 725, a chimeric gene. However, this gene was not detected in onion accessions containing CMS-T cytoplasm. Instead, a region showing high homology with orfA501, which is located in the 3’ end of orf725, was detected in CMS-T. A novel chimeric gene, which we named orf219, was identified by genome walking PCR amplification based on the orfA501 homolog. The 684-bp open reading frame of orf219 consisted of an 128-bp of exon 1 of atp1 and a 556-bp sequence of an orfA501 homolog. The high-copy-number orf219 was detected only in the CMS-T cytoplasm (T cytotype). Analysis of RT-PCR products showed normal transcription of orf219 and eight RNA editing sites, one of which created a stop codon, resulting in a shorter amino acid sequence upon translation. Sequences of four hypervariable regions in the chloroplast genome and the organization of syntenic blocks in mitochondrial genome indicated that CMS-T was very closely related to the normal (N) male-fertile and CMS-R cytotypes. Based on these findings, new molecular markers were developed for the identification of cytotypes CMS-T, -R, -S, and -N. Cytotypes of 424 diverse onion accessions were identified using six molecular markers. The CMS-S and CMS-R cytotypes predominated among the analyzed accessions. In contrast, only five accessions possessed CMS-T, supporting earlier reports that CMS-T is rarely used in hybrid-onion breeding.





Similar content being viewed by others
References
Abdelnoor RV, Christensen AC, Mohammed S, Munoz-Castillo B, Moriyama H, Mackenzie SA (2006) Mitochondrial genome dynamics in plants and animals: convergent gene fusions of a MutS homologue. J Mol Evol 63:165–173
Albert B, Godelle B, Gouyon PH (1998) Evolution of the plant mitochondrial genome: dynamics of duplication and deletion of sequences. J Mol Evol 46:155–158
Alcalá J, Pike LM, Giovannoni JJ (1999) Identification of plastome variants useful for cytoplasmic selection and cultivar identification in onion. J Am Soc Hort Sci 124:122–127
Allen JO, Fauron CM, Mink P, Roark L, Oddiraju S, Lin GN, Meyer L, Sun H, Kim K, Wang C, Du F, Xu D, Gibson M, Cifrese J, Clifton SW, Newton KJ (2007) Comparisons among two fertile and three male-sterile mitochondrial genomes of maize. Genetics 177:1173–1192
Arrieta-Montiel M, Lyznik A, Woloszynska M, Janska H, Tohme J, Mackenzie SA (2001) Tracing evolutionary and developmental implications of mitochondrial stoichiometric shifting in the common bean. Genetics 158:851–864
Backert S, Neilsen BL, Börner T (1997) The mystery of the rings: structure and replication of mitochondrial genomes from higher plants. Trends Plant Sci 2:477–483
Bellaoui M, Martin-Canadell A, Pelletier G, Budar F (1998) Low-copy-number molecules are produced by recombination, actively maintained and can be amplified in the mitochondrial genome of Brassicaceae: relationship to reversion of the male sterile phenotype in some cybrids. Mol Gen Genet 257:177–185
Berninger E (1965) Contribution à l’étude de la sterilité mâle de l’oignon (Allium cepa L.). Ann Amélior Plant 15:183–199
Bohra A, Jha UC, Adhimoolam P, Bisht D, Singh NP (2016) Cytoplasmic male sterility (CMS) in hybrid breeding in field crops. Plant Cell Rep 35:967–993
Budar F, Touzet P, De Paepe R (2003) The nucleo-mitochondrial conflict in cytoplasmic male sterilities revised. Genetica 117:3–16
Chen L, Liu Y (2013) Male sterility and fertility restoration in crops. Ann Rev Plant Biol 65:579–606
Chen R, Zhao N, Li S, Grover CE, Nie H, Wendel JF, Hua J (2017) Plant mitochondrial genome evolution and cytoplasmic male sterility. Crit Rev Plant Sci 36:55–69
Cui X, Wise RP, Schnable PS (1996) The rf2 nuclear restorer gene of male-sterile T-cytoplasm maize. Science 272:1334–1336
Doyle JJ, Doyle JL (1987) A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull 19:11–15
Engelke T, Terefe D, Tatlioglu T (2003) A PCR-based marker system monitoring CMS-(S), CMS-(T) and (N)-cytoplasm in the onion (Allium cepa L.). Theor Appl Genet 107:162–167
EngelkeTatlioglu T (2002) A PCR-marker for the CMS1 inducing cytoplasm in chives derived from recombination events affecting the mitochondrial gene atp9. Theor Appl Genet 104:698–702
Fauron CMR, Moore B, Casper M (1995) Maize as a model of higher plant mitochondrial genome plasticity. Plant Sci 112:11–32
Fujii S, Toriyama K (2009) Suppressed expression of retrograde-regulated male sterility restores pollen fertility in cytoplasmic male sterile rice plants. Proc Natl Acad Sci USA 106:9513–9518
Gaborieau L, Brown GG, Mireau H (2016) The propensity of pentatricopeptide repeat genes to evolve into restorers of cytoplasmic male sterility. Front Plant Sci 7:1816
Hanson MR, Bentolila S (2004) Interactions of mitochondrial and nuclear genes that affect male gametophyte development. Plant Cell 16:S154–S169
Havey MJ (1993) A putative donor of S-cytoplasm and its distribution among open-pollinated populations of onion. Theor Appl Genet 86:128–134
Havey MJ (1995) Identification of cytoplasms using the polymerase chain reaction to aid in the extraction of maintainer lines from open-pollinated populations of onion. Theor Appl Genet 90:263–268
Havey MJ (2000) Diversity among male-sterility-inducing and male-fertile cytoplasms of onion. Theor Appl Genet 101:778–782
Havey MJ, Kim S (2021) Molecular marker characterization of commercially used cytoplasmic male sterilities in onion. J Amer Soc Hort Sci 146:351–355
Hu J, Wang K, Huang W, Liu G, Gao Y, Wang J, Huang Q, Ji Y, Qin X, Wan L, Zhu R, Li S, Yang D, Zhu Y (2012) The rice pentatricopeptide repeat protein RF5 restores fertility in Hong–Lian cytoplasmic male-sterile lines via a complex with the glycine-rich protein GRP162. Plant Cell 24:109–122
Janska H, Sarria R, Woloszynska M, Arrieta-Montiel M, Mackenzie SA (1998) Stoichiometric shifts in the common bean mitochondrial genome leading to male sterility and spontaneous reversion to fertility. Plant Cell 10:1163–1180
Jones HA, Clarke A (1943) Inheritance of male sterility in the onion and the production of hybrid seed. Proc Am Soc Hort Sci 43:189–194
Jones HA, Emsweller SL (1936) A male-sterile onion. Proc Am Soc Hort Sci 34:582–585
Kim S (2013) Identification of hypervariable chloroplast intergenic sequences in onion (Allium cepa L.) and their use to analyse the origins of male-sterile onion cytotypes. J Hort Sci Biotechnol 88:187–194
Kim S (2014) A codominant molecular marker in linkage disequilibrium with a restorer-of-fertility gene (Ms) and its application in reevaluation of inheritance of fertility restoration in onions. Mol Breed 34:769–778
Kim B, Kim S (2019) Identification of a variant of CMS-T cytoplasm and development of high resolution melting markers for distinguishing cytoplasm types and genotyping a restorer-of-fertility locus in onion (Allium cepa L.). Euphytica 215:164
Kim S, Yoon M (2010) Comparison of mitochondrial and chloroplast genome segments from three onion (Allium cepa L.) cytoplasm types and identification of a trans-splicing intron of cox2. Curr Genet 56:177–188
Kim Y, Zhang D (2018) Molecular control of male fertility for crop hybrid breeding. Trends Plant Sci 23:53–65
Kim S, Lee E, Cho DY, Han T, Bang H, Patil BS, Ahn YK, Yoon M (2009a) Identification of a novel chimeric gene, orf725, and its use in development of a molecular marker for distinguishing three cytoplasm types in onion (Allium cepa L.). Theor Appl Genet 118:433–441
Kim S, Lee E, Kim C, Yoon M (2009b) Distribution of three cytoplasm types in onion (Allium cepa L.) cultivars bred in Korea and Japan. Kor J Hort Sci Technol 27:275–279
Kim S, Bang H, Patil BS (2013) Origin of three characteristic onion (Allium cepa L.) mitochondrial genome rearrangements in Allium species. Sci Hortic 157:24–31
Kim S, Kim C, Park M, Choi D (2015a) Identification of candidate genes associated with fertility restoration of cytoplasmic male sterility in onion (Allium cepa L.) using a combination of bulked segregant analysis and RNA-seq. Theor Apple Genet 128:2289–2299
Kim S, Park J, Yang T (2015b) Characterization of three active transposable elements recently inserted in three independent DFR-A alleles and one high-copy DNA transposon isolated from the Pink allele of the ANS gene in onion (Allium cepa L.). Mol Genet Genomics 290:1027–1037
Kim S, Park J, Yang T (2015c) Comparative analysis of the complete chloroplast genome sequences of a normal male-fertile cytoplasm and two different cytoplasms conferring cytoplasmic male sterility in onion (Allium cepa L.). J Hort Sci Biotechnol 90:459–468
Kim B, Kim K, Yang T, Kim S (2016) Completion of the mitochondrial genome sequence of onion (Allium cepa L.) containing the CMS-S male-sterile cytoplasm and identification of an independent event of the ccmFN gene split. Curr Genet 62:873–885
Kim B, Kim C, Kim S (2019a) Inheritance of fertility restoration of male sterility conferred by cytotype Y and identification of instability of male fertility phenotypes in onion (Allium cepa L.). J Hort Sci Biotechnol 94:341–348
Kim B, Yang T, Kim S (2019b) Identification of a gene responsible for cytoplasmic male sterility in onions (Allium cepa L.) using comparative analysis of mitochondrial genome sequences of two recently diverged cytoplasms. Theor Appl Genet 132:313–322
Kitazaki K, Kubo T (2010) Cost of having the largest mitochondrial genome: Evolutionary mechanism of plant mitochondrial genome. J Bot 2010:620137
Kitazaki K, Arakawa T, Matsunaga M, Yui-Kurino R, Matsuhira H, Mikami T, Kubo T (2015) Post-translational mechanisms are associated with fertility restoration of cytoplasmic male sterility in sugar beet (Beta vulgaris). Plant J 83:290–299
Kmiec B, Woloszynska M, Janska H (2006) Heteroplasmy as a common state of mitochondrial genetic information in plants and animals. Curr Genet 50:149–159
McCauley DE, Bailey MF (2009) Recent advances in the study of gynodioecy: the interface of theory and empiricism. Ann Bot 104:611–620
Oldenburg DJ, Bendich AJ (2001) Mitochondrial DNA from the Liverwort Marchantia polymorpha: Circularly permuted linear molecules, head-to-tail concatemers, and a 5’ protein. J Mol Biol 310:549–562
Sakai T, Imamura J (1993) Evidence for a mitochondrial sub-genome containing radish AtpA in a Brassica napus cybrid. Plant Sci 90:95–103
Sandhu AP, Abdelnoor RV, Mackenzie SA (2007) Transgenic induction of mitochondrial rearrangements for cytoplasmic male sterility in crop plants. Proc Natl Acad Sci USA 104:1766–1770
Satoh M, Kubo T, Nishizawa S, Estiati A, Itchoda N, Mikami T (2004) The cytoplasmic male-sterile type and normal type mitochondrial genomes of sugar beet share the same complement of genes of known function but differ in the content of expressed ORFs. Mol Genet Genomics 272:247–256
Schnable PS, Wise RP (1998) The molecular basis of cytoplasmic male sterility and fertility restoration. Trends Plant Sci 3:175–180
Schweisguth B (1973) Étude d’un nouveau type de stérilité male chez l’oignon, Allium cepa L. Ann Amélior Plant 23:221–233
Skippington E, Barkman TJ, Rice DW, Palmer JD (2015) Miniaturized mitogenome of the parasitic plant Viscum scurruloideum is extremely divergent and dynamic and has lost all nad genes. Proc Natl Acad Sci USA 112:E3515-3524
Sloan DB (2013) One ring to rule them all? Genome sequencing provides new insights into the “master circle” model of plant mitochondrial DNA structure. New Phytol 200:978–985
Sloan DB, Alverson AJ, Chuckalovcak JP, Wu M, McCauley DE, Palmer JD, Taylor DR (2012) Rapid evolution of enormous, multichromosomal genomes in flowering plant mitochondria with exceptionally high mutation rates. PLoS Biol 10:e1001241
Small I, Suffolk R, Leaver CJ (1989) Evolution of plant mitochondrial genomes via substoichiometric intermediates. Cell 58:69–76
Tsujimura M, Kaneko T, Sakamoto T, Kimura S, Shigyo M, Yamagishi H, Terachi T (2019) Multichromosomal structure of the onion mitochondrial genome and a transcript analysis. Mitochondrion 46:179–186
Woloszynska M, Trojanowski D (2009) Counting mtDNA molecules in Phaseolus vulgaaris: sublimons are constantly produced by recombination via short repeats and undergo rigorous selection during substoichiometric shifting. Plant Mol Biol 70:511–521
Yu N, Kim S (2021) Identification of Ms2, a novel locus controlling male-fertility restoration of cytoplasmic male sterility in onion (Allium cepa L.), and development of tightly linked molecular markers. Euphytica 217:191
Acknowledgements
The authors thank Ji-hwa Heo, Jeong-An Yoo, and Su-jeong Kim for their technical assistance and Dr. Michael Havey for DNA of RJ70A, the source of CMS-T.
Funding
This study was supported by the Korea Institute of Planning and Evaluation for Technology in Food, Agriculture and Forestry (No. 322067-03-1-HD020) and the BK21 FOUR funded by the Ministry of Education of Korea and National Research Foundation of Korea.
Author information
Authors and Affiliations
Contributions
WA: investigation, methodology, data curation, writing—original draft; SK: conceptualization, investigation, methodology, writing—review and editing, supervision.
Corresponding author
Ethics declarations
Conflict of interests
The authors declare no competing interests.
Ethics approval
All experiments were performed in compliance with the current laws of the Republic of Korea.
Consent for publication
Not applicable.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Ahn, W., Kim, S. Identification of a candidate gene responsible for male sterility conferred by CMS-T cytoplasm in onion (Allium cepa L.) and development of molecular markers for detection of CMS-T cytoplasm. Euphytica 219, 28 (2023). https://doi.org/10.1007/s10681-023-03158-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s10681-023-03158-5