Abstract
Reniform nematode (Rotylenchulus reniformis, Linford and Oliveira) is a sedentary, semi-endoparasite that infects a wide range of plant hosts and is one of the top three nematode pathogens affecting soybean in the southeastern United States. Previous studies have linked resistance to reniform nematode in soybean to two quantitative trait loci on chromosomes 11 and 18. A Kompetitive Allele-Specific PCR (KASP) assay was designed using SNP markers within these two regions to distinguish reniform nematode-resistant soybean based on genotype. A collection of 44 soybean plant introductions with resistant phenotype to reniform nematode and 40 susceptible soybean lines were genotyped at the two target loci to validate the KASP assay design. Of the 44 observed resistant lines, two carried the susceptible genotype; PI 438489B at the locus on chromosome 18 and PI 495017C on chromosome 11. Of the 40 observed susceptible soybean lines, only 25 had the expected susceptible genotype at the loci on chromosome 18 and 13 on chromosome 11. Our KASP assay was 68% accurate in predicting the phenotype of 84 soybean accessions based on their genotype at the SNP marker on chromosome 18 and 83% accurate at chromosome 11. These results indicate a moderate correlation of soybean SNP markers GlyREN18_46 and GlyREN11_190 with reniform nematode resistance. Further research is required to improve the accuracy of KASP assays to predict soybean response to reniform nematode, particularly host susceptibility.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Reniform nematode (Rotylenchulus reniformis, Linford and Oliveira) is a major yield-limiting pathogen of soybean throughout most of the southeastern United States, responsible for 11–33% of the yield loss (Allen et al. 2021; Rebois and Johnson 1973; Robbins et al. 1994a). The use of resistant cultivars is the best management practice to prevent yield loss and reduce nematode populations in the soil if nematode populations are above damage thresholds (Westphal and Scott 2005). Current nematode-resistant cultivars of soybean are tailored to protect against soybean cyst nematode (SCN) which is controlled in part by the major soybean loci Rhg1 and Rhg4 on chromosomes (Chrs.) 18 and 8, respectively (Liu et al. 2017). A combination of both rhg1-a and rhg4 alleles have been shown to carry resistance to reniform nematode (RN) and SCN, but some SCN-resistant genotypes contain only the rhg1-b allele which is not linked to RN resistance (Cook et al. 2012; Klepadlo et al. 2018). Unfortunately, many of the SCN-resistant soybean cultivars available for commercial production are derived from either Plant Introduction (PI) 88788 or Peking, which do not carry the resistant allele at Rhg4 and are therefore susceptible to RN (McCarville et al. 2017).
Studies over the last 20 years have investigated the performance of hundreds of released cultivars and breeding lines of soybean against RN (Lee et al. 2015; Robbins and Rakes 1996; Robbins et al. 1994b, 1999, 2001, 2002, 2017; Stetina et al. 2014). These studies highlight the wide spectrum of host responses to RN in PI lines and cultivars and identify lines that carry genetic resistance to this nematode pathogen. Manual screening for resistance, such as quantifying the reproduction index (RI) in large populations of breeding lines, requires a high cost, large space, and months of labor (Jenkins 1964; Robbins and Rakes 1996). With the advent of molecular tools, breeders have the ability to test their breeding populations for desired genetic characteristics, such as disease resistance, within a fraction of the time and cost compared to manual screening.
Several rapid molecular screening techniques have been developed to assist breeders in predicting desired phenotypes using genomic markers (Broccanello et al. 2018; Semagn et al. 2014). A KASP (Kompetitive Allele-Specific PCR) assay is one of these marker detection assays that is rapid, reliable, and has a low cost per sample. This PCR-based assay targets single nucleotide polymorphisms (SNPs) through oligo extension and uses fluorescence resonance energy transfer (FRET) for signal generation (Kumpatla et al. 2012). KASP assays are best suited for studies targeting between one and 10 SNP markers in hundreds of plant accessions, which is often the case when screening for markers linked to desired traits in large breeding populations. Although inheritance of qualitative traits such as pathogen resistance is complex, assays designed for marker-assisted selection have improved efforts to pyramid desired traits and increase the efficiency of breeding programs (Semagn et al. 2014). For example, a KASP assay designed to target three SNP markers has proven highly effective in identifying SCN resistance (Shi et al. 2015).
Recent studies have linked two QTL regions to RN resistance on soybean Chrs. 18 and 11. One study identified a region on Chr. 19, yet no subsequent study has associated the resistance phenotype to a genomic region on this chromosome (Ha et al. 2007; Jiao et al. 2015; Lee et al. 2016; Wilkes et al. 2020). A recent study by Klepadlo et al. (2018) screened 76 SCN-resistant soybean accessions for RN resistance using both phenotypic observations and genotypic screening using a KASP assay. In this study, a developed KASP marker on Chr. 18 had a 90% correlation with the expected phenotype, but there was low phenotype correlation with the markers developed on Chrs. 11 and 19. Further research is needed to survey a broader genetic pool, including accessions susceptible to SCN, to provide a more comprehensive understanding of the genetic regions associated with RN resistance.
This study aims to develop a KASP assay to assist in the marker-assisted selection of RN resistance in soybean. The SNP markers within two QTL regions, which accounted for 44% of the total phenotypic variance, were used to design the genotyping assay (Wilkes et al. 2020). Forty-four RN-resistant and 40 RN-susceptible soybean lines (84 total) were genotyped using the assay to test accuracy. Additionally, the phenotypic responses of the 44 RN-resistant soybean lines were assessed with a RN population from South Carolina.
Materials and methods
Manual reniform nematode resistance screening
All soybean accessions reported to be resistant to RN by the Germplasm Resources Information Network (GRIN, USDA-ARS) were acquired (44 accessions) and screened for resistance to a RN population collected in South Carolina. The soybeans were planted in 20-ounce Styrofoam cups in three replicates and grown in a temperature-controlled growth room. The room was maintained at 38 °C and pots were watered roughly 50 mL once a day using an automated irrigation system. A positive control (RN-susceptible soybean cultivar Braxton) was planted in three replicates in conjunction with each trial. Once the first trifoliate emerged, six days after planting, each plant was individually inoculated with 2000 vermiform RN. The RN population was originally collected from St. Matthews, South Carolina and cultured for more than 30 life cycles on cv. Braxton in a controlled environment. After 30 days post-inoculation, all soybean plants were harvested and nematodes were extracted from the soil in each pot (Jenkins 1964). The RI was calculated to estimate the nematode reproduction on each soybean accession (Perry et al. 2018).
DNA extraction
Forty RN-susceptible soybean lines were randomly selected from previous RN screening projects with an emphasis to sample lines with no overlapping pedigrees (Lee et al. 2015; Robbins et al. 2017). Each accession was planted in the same growth room with the resistant accessions. A penny-sized amount of newly emerging plant leaf material was collected from all soybean accessions, both RN-resistant and susceptible, for DNA extraction using a sodium dodecyl sulfate (SDS) protocol published by King et al. (2014). Each sample of extracted DNA was normalized to a concentration of 50 ng/µL for use in the KASP assay.
Primer design
Two primer sets were designed to target SNPs within QTL regions previously identified in Wilkes et al. (2020) (Table 1). The first target SNP, referred to as GlyREN18_46, was on Chr. 18 at bp 1684449. At this marker, the resistant genotype was C and the susceptible genotype was A. The second target SNP was on Chr. 11 at bp 32986440, referred to as GlyREN11_190. The resistant genotype for GlyREN11_190 was C and the susceptible genotype was T. The 5’ end of each forward primer was appended with a complementary sequence to the FAM fluorophore for the susceptible genotype from cv. Williams 82 and HEX fluorophore quenchers for the resistant genotype from cv. Forrest. Quality of the primer design was reviewed using the OligoAnalyzer Tool from IDT (idtdna.com). A BLAST search on the Soybase Database using the Glycine max reference genome (Wm81.a4) revealed that neither primer set produced an off-target 100% matched hit.
KASP assay
A modified KASP assay protocol was used (Patterson et al. 2017). Stock primers were resuspended in ddH2O at a concentration of 100 µM. A primer master mix was formulated based on with the following: 18 µL of forward primer with FAM sequence, 18 µL of forward primer with HEX sequence, 45 µL of the reverse primer, and 69 µL of 1 M Tris-HCl, pH 8.3. A KASP master mix for each target SNP was made at a ratio of 514 µL of the KASP V4.0 2X MasterMix (LGC Genomics™) combined with 14 µL of the primer master mix previously described.
A 96-well plate was prepared by pipetting 4 µl of the KASP master mix followed by 4 µl of DNA sample or ddH2O for the non-template control (NTC). Standard controls for each genotype were replicated at least three times and each unknown sample had at least two replicates per run. Soybean cultivars Forrest (resistant) and Williams 82 (susceptible) were used as the homozygous standards. Heterozygous standards were selected from a set of recombinant inbred lines developed from a cross between these two controls (Wilkes et al. 2020; Wu et al. 2011). Plates were sealed with optically clear adhesive plate seals (Thermo Scientific Cat. No. AB1170). The assay was performed using a Bio-Rad CFX Connect Real-Time PCR Detection System (Bio-Rad Laboratories, Inc., Hercules, CA) using a modified LGC qPCR program: 10 cycles of 94 °C for 15 min, 94 °C for 20 s, 61–55 °C for 60 s, dropping 0.6 °C per cycle followed by 30 cycles of DNA separation at 94 °C for 20 s, annealing at 55 °C for 1 min, and elongation at 23 °C for 30 s. HEX and FAM fluorescence strength was measured at the end of each of the final 30 DNA amplification cycles.
Data analysis
FAM and HEX fluorescence values were normalized by transforming each value to a percentage of the maximum fluorescence for each fluorophore in each assay (Patterson et al. 2017). A discriminate analysis was performed to identify statistically distinct clusters of similar HEX and FAM fluorescence at each target site to call genotype (JMP® 2020, SAS institute).
Results
Manual reniform nematode resistance screening
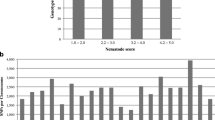
All 44 GRIN-labeled RN-resistant soybean lines had lower RI compared to the susceptible control, cv. Braxton. However, six of the 44 lines had a slightly higher RI than the other 38 tested lines (Fig. 1). These genotypes were PI 438489B, PI 495017C, PI 467332, PI 467312, PI 458520, and PI 416762. An optimal univariate cluster analysis was performed on the RI of all phenotyped soybeans to clearly illustrate the distinct grouping of these six individuals apart from the other accessions (Fig. 2) (Usovsky et al. 2022).
Reproduction Indices (RI) of 44 diverse reniform nematode-resistant soybean germplasm as indicated by the Germplasm Resources Information Network (GRIN, USDA-ARS). Soybean cultivar, Braxton, was used as a susceptible control and an average of 3 replicates was used in each trial. Blue bar represents trial 1, red trial 2, and green trial 3
KASP assay
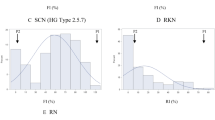
The KASP assay designed to target SNP alleles at GlyREN18_46 and GlyREN11_190 successfully distinguished the genotypes between the susceptible and resistant standard controls in all the trials. However, at marker GlyREN11_190, the homozygous susceptible standard had higher HEX fluorescence than expected and was indistinguishable from the heterozygous standard. The resistant controls remained distinct from the susceptible and heterozygous standards with high HEX signal and low to no FAM signal.
Of the 44 RN-resistant soybean lines, two accessions had genotype calls consistent with the susceptible standard (Fig. 3). One accession (PI 438489B) had the susceptible genotype at GlyREN18_46 and resistant genotype at GlyREN11_190. The other accession (PI 495017C) had the susceptible genotype at the GlyREN11_190 and resistant genotype at GlyREN18_46. These two unique accessions are two of the six resistant accessions that were phenotypically distinct based on the cluster analysis of RI values. The other four of the six accessions had the expected resistant genotype at both loci.
The probability of an accurate correlation of genotype with phenotype in the observed RN-resistant accessions at GlyREN18_46 was estimated at 97.7%. However, the results from the RN-susceptible soybean accessions at both markers were less accurately correlated with the observed phenotype (Fig. 4). For marker GlyREN18_46, 14 RN-susceptible soybean lines had a genotype consistent with the susceptible standard and 25 RN-susceptible lines had a genotype consistent with the resistant standard (one accession had no amplification). For GlyREN11_190, 26 RN-susceptible soybean accessions had a genotype consistent with the susceptible standard, 13 accessions had a genotype consistent with the resistant standard, and one accession (SC06-306) had FAM and HEX fluorescence signals distinct from all other data points (Fig. 4). Results from genotyping the RN-susceptible soybean accessions show a 36 and 66.7% phenotype prediction accuracy for SNP alleles at GlyREN18_46 and GlyREN11_190, respectively. The combined results of the RN-susceptible and resistant accessions show genotyping at GlyREN18_46 and GlyREN_11_190 was 68% and 83% accurate, respectively, in predicting RN response in soybean.
Discussion
The 44 GRIN-labeled RN-resistant soybean accessions had reduced RN reproduction compared to a susceptible control when screened with an RN population originally collected from South Carolina. However, six of the 44 RN-resistant soybean lines had consistently higher RI than the other 38 resistant lines (Fig. 2). Three (PI 467312, PI 458520, PI 416762) of those six were once reported susceptible to RN (Lee et al. 2016). Similarly, two of the same lines (PI 458520, PI 416762) were labeled susceptible in a separate study (Klepadlo et al. 2018; Lee et al. 2016). Additionally, PI 437725 had low RI, consistent with the highly resistant accessions but was classified as susceptible in the Lee et al. (2016) report. One possible explanation for the inconsistency is host response to a RN population from a different geographic region. Our study uses a RN population from South Carolina while most other reports used a RN population from Arkansas. Differences in environmental conditions of the assay, such as temperature of the soil, size of the pot, and moisture content, may also affect the host response to the pathogen. The seed source may have resulted in an unexpected response to RN. Additionally, there may be small-effect QTLs that are part of the overall phenotype. The six accessions with distinct RI values (Fig. 2) may highlight a group of accessions that have similar genetic sources of resistance which include small effect QTLs.
The KASP assay results show that two of the genotyped RN-resistant soybean lines carried the susceptible genotype: PI 438489B at GlyREN18_46 and PI 495017C at GlyREN11_190. These soybean lines were two of the six accessions with slightly higher RI than the other 38 RN-resistant accessions. It is possible that PI 438489B and PI 495017C may have a different source of genetic resistance that accounts for the susceptible genotypes at the two loci and a slightly higher nematode reproduction than other resistant accessions. It would be of interest to investigate these six soybean lines, particularly the two with different genotypes, to identify novel major and minor QTLs.
Measuring phenotypic resistance to a nematode pathogen is not a straightforward process. Resistance to reniform nematode is most often represented by the RI, a relative measurement of a host’s suitability to the pathogen. These values are on a continuous spectrum which makes classifying resistance a challenge. Additionally, the RI of an individual plant can be affected by environmental conditions, such as temperature and soil texture, which is the reason many studies use a standardized control for relative comparison (Perry et al. 2018). However, comparisons to the control can lead to inconsistent host suitability calls since it is not uncommon for the RI distribution in a population to fall outside the range of the controls. This further justifies the need for a reliable genotyping method to define and classify nematode resistance.
The KASP assay designed to target GlyREN18_46 gave three distinct clusters (homozygous resistant, homozygous susceptible, and heterozygous) among tested soybean accessions (Figs. 3, 4). However, the assay design targeting GlyREN11_190 did not provide fluorescence measurements in three clusters as expected. The resistant standard gave the expected result of a high HEX fluorescent signal and low FAM signal, indicating a cluster with the TT genotype. The heterozygous standard also gave expected results of equal signals of both FAM and HEX indicating a cluster with CT genotype. However, the susceptible control aligned with the heterozygous standard, with higher than expected HEX signal. Likewise, several susceptible soybean accessions had genotypes that aligned with the heterozygous and homozygous controls. Interestingly, one susceptible line, SC06-306, had unique fluorescence signals apart from the other accessions with a higher HEX signal (Fig. 4b). In-depth research, such as amplicon sequencing of each genotype, would provide a clear understanding of the genotype of this locus among the sample population.
Although the marker at GlyREN11_190 did not perform as expected in the assay, there was a clear distinction between resistant and susceptible standard controls. Conclusions can be drawn from results of the genotyped soybean lines to indicate the use for genotype to phenotype correlation. The accuracy rate for predicting the phenotypically resistant soybean accessions was higher than the rate for the susceptible accessions. The RN-susceptible accessions may have greater genetic diversity compared to resistant genotypes. The resistant soybean lines may come from similar pedigrees, particularly if there was one original source of resistance, and therefore have more similar haplotypes. Additionally, being a quantitative trait, there are multiple loci involved in the resistance phenotype. Testing for one or two known RN resistance loci may not be sufficient to predict the phenotype and therefore lead to a lower prediction accuracy.
Klepadlo et al. (2018) developed three KASP markers to test for RN resistance and found an overall correlation of 67.2% linking the genotype with the phenotypic variation in 76 soybean accessions. Our study screened 84 soybean lines and shows a comparable rate of 76%, accurately correlating genotype to phenotype with only two markers. Specifically, the accuracy of GlyREN11_190 was 83% compared to Klepadlo’s 63% from a maker within the same QTL region. However, the marker at GlyREN18_46 was much lower in accuracy compared to their reported 89%. The marker GlyREN18_46 was 666 kb downstream on Chr. 18 of the KASP marker developed by Klepadlo et al. and GlyREN11_190 was 27 kbp downstream from their marker on Chr. 11. A combination of the KASP-designed marker on Chr. 18 in the study done by Klepadlo et al. and GlyREN11_190 would provide an even greater accuracy in phenotype prediction.
Our study contributes to the ongoing efforts to design and implement a fast molecular assay for soybean targeting known loci associated with resistance to RN. Development of a quick and reliable genetic screening for RN resistance in soybean can assist breeders in the rapid selection of genotypes with known nematode resistance.
References
Allen TW, Bissonnette K, Bradley CA, Damicone JP, Dufault NS, Faske TR, Isakeit T, Kemerait RC, Koehler A, Mehl HL, Mueller JD, Padgett GB, Price PP, Sikora EJ, Small IM, Thiessen L, Young H (2021) Southern United States soybean disease loss estimates for 2021. Proceedings of the 46th Annual Meeting of the Southern Soybean Disease Workers. Pensacola FL
Broccanello C, Chiodi C, Funk A, McGrath JM, Panella L, Stevanato P (2018) Comparison of three PCR-based assays for SNP genotyping in plants. Plant Methods 14:28
Cook DE, Lee TG, Guo X, Melito S, Wang K, Bayless AM, Wang J, Hughes TJ, Willis DK, Clemente TE et al (2012) Copy number variation of multiple genes at Rhg1 mediates nematode resistance in soybean. Science 338:1206–1209
Ha B-K, Robbins RT, Han F, Hussey RS, Soper JF, Boerma HR (2007) SSR mapping and confirmation of soybean QTL from PI 437654 conditioning resistance to reniform nematode. Crop Sci 47:1336–1343
Jenkins WR (1964) A rapid centrifugal-flotation technique for separating nematodes from soil. Plant Dis Rep 48:692
Jiao Y, Vuong TD, Liu Y, Li Z, Noe J, Robbins RT, Joshi T, Xu D, Shannon JG, Nguyen HT (2015) Identification of quantitative trait loci underlying resistance to southern root-knot and reniform nematodes in soybean accession PI 567516 C. Mol Breed 35:131
King Z, Serrano J, Boerma HR, Li Z (2014) Non-toxic and efficient DNA extractions for soybean leaf and seed chips for high-throughput and large-scale genotyping. Biotechnol Lett 36:1875–1879
Klepadlo M, Meinhardt CG, Vuong TD, Patil G, Bachleda N, Ye H, Robbins RT, Li Z, Shannon JG, Chen P et al (2018) Evaluation of soybean germplasm for resistance to multiple nematode species: Heterodera glycines, Meloidogyne incognita, and Rotylenchulus reniformis. Crop Sci 58:2511–2522
Kumpatla SP, Buyyarapu R, Abdurakhmonov IY, Mammadov JA (2012) Genomics-assisted plant breeding in the 21st century: technological advances and progress. IntechOpen
Lee J-D, Kim HJ, Robbins RT, Wrather JA, Bond J, Nguyen HT, Shannon JG (2015) Reaction of soybean cyst nematode resistant plant introductions to root-knot and reniform nematodes. Plant Breed Biotechnol 3:346–354
Lee Y-C, Lightfoot DA, Anderson J, Robbins RT, Kantartzi SK (2016) QTL underlying reniform nematode resistance in soybean cultivar Hartwig. Atlas J Biol 2016:308–312
Liu S, Kandoth PK, Lakhssassi N, Kang J, Colantonio V, Heinz R, Yeckel G, Zhou Z, Bekal S, Dapprich J et al (2017) The soybean GmSNAP18 gene underlies two types of resistance to soybean cyst nematode. Nat Commun 8:14822
McCarville MT, Marett CC, Mullaney MP, Gebhart GD, Tylka GL (2017) Increase in soybean cyst nematode virulence and reproduction on resistant soybean varieties in Iowa from 2001 to 2005 and the effects on soybean yields. Plant Health Progress 18(3):146–155
Patterson EL, Fleming MB, Kessler KC, Nissen SJ, Gaines TA (2017) A KASP genotyping method to identify northern watermilfoil, Eurasian watermilfoil, and their interspecific hybrids. Front Plant Sci 8:752
Perry RN, Moens M, Jones JT (2018) Cyst nematodes-Life cycle and economic importance Cyst nematodes. CABI, Wallingford, UK, pp 1–26
Rebois RV, Johnson WC (1973) Effect of Rotylenchulus reniformis on yield and nitrogen, potassium, phosphorus and amino acid content of seed of Glycine max. J Nematol 5:1–6
Robbins RT, Rakes L (1996) Resistance to the reniform nematode in selected soybean cultivars and germplasm lines. J Nematol 28:612–615
Robbins RT, Rakes L, Elkins CR (1994a) Reniform nematode reproduction and soybean yield of four soybean cultivars in Arkansas. J Nematol 26:656–658
Robbins RT, Rakes L, Elkins CR (1994b) Reproduction of the reniform nematode on thirty soybean cultivars. J Nematol 26:659–664
Robbins RT, Rakes L, Jackson LE, Dombek DG (1999) Reniform nematode resistance in selected soybean cultivars. J Nematol 31:667–677
Robbins RT, Rakes L, Jackson LE, Gbur EE, Dombek DG (2001) Host suitability in soybean cultivars for the reniform nematode, 2000 tests. J Nematol 33:314–317
Robbins RT, Shipe ER, Rakes L, Jackson LE, Gbur EE, Dombek DG (2002) Host suitability of soybean cultivars and breeding lines to reniform nematode in tests conducted in 2001. J Nematol 34:378–383
Robbins RT, Arelli P, Chen P, Shannon G, Kantartzi S, Fallen B, Li Z, Faske T, Velie J, Gbur E et al (2017) Reniform nematode reproduction on soybean cultivars and breeding lines in 2016. Proceeding 2016 Beltwide cotton conference. January 4–6 2017 Dallas TX USA184–214
Semagn K, Babu R, Hearne S, Olsen M (2014) Single nucleotide polymorphism genotyping using Kompetitive Allele specific PCR (KASP): overview of the technology and its application in crop improvement. Mol Breed 33:1–14
Shi Z, Liu S, Noe J, Arelli P, Meksem K, Li Z (2015) SNP identification and marker assay development for high-throughput selection of soybean cyst nematode resistance. BMC Genomics 16:314
Stetina SR, Smith JR, Ray JD (2014) Identification of Rotylenchulus reniformis resistant Glycine lines. J Nematol 46:1–7
Usovsky M, Robbins RT, Wilkes JF, Crippen D, Shankar D, Vuong TD, Agudelo P, Nguyen HT (2022) Classification methods and identification of reniform nematode resistance in known soybean cyst nematode resistant soybean genotypes. Plant Dis 106(2):382–389. https://doi.org/10.1094/PDIS-01-21-0051-RE
Westphal A, Scott AW (2005) Implementation of soybean in cotton cropping sequences for management of reniform nematode in south Texas. Crop Sci 45:233–239
Wilkes J, Saski CA, Klepadlo M, Fallen B, Agudelo P (2020) Quantitative trait loci associated with Rotylenchulus reniformis host suitability in soybean. Phytopathology 110(9):1511–1521. https://doi.org/10.1094/PHYTO-02-20-0035-R
Wu X, Vuong TD, Leroy JA, Grover SJ, Sleeper DA, Nguyen HT (2011) Selection of a core set of RILs from Forrest x Williams 82 to develop a framework map in soybean. Theor Appl Genet 122(6):1179–1187
Funding
The authors would like to thank the South Carolina Soybean Board for partial funding of this research. Mention of trade names or commercial products in this publication is solely for the purposes of providing information and does not imply recommendations or endorsement by the USDA. USDA is an equal opportunity provider and employer.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflicts of interest.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Wilkes, J.E., Fallen, B., Saski, C. et al. Development of SNP molecular markers associated with resistance to reniform nematode in soybean using KASP genotyping. Euphytica 219, 27 (2023). https://doi.org/10.1007/s10681-022-03144-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s10681-022-03144-3