Abstract
The peanut rich in oleic acid in seed oil, also known as high oleic peanut, renders its products superior in human healthy diet, long shelf life and high oxidative stability in food and oil processing. High oleic peanut varieties in China are derived from a handful of high oleic donors with limited genetic background. Improvement of high oleic peanut cultivars by integrating key target traits is crucial for both food security and germplasm enhancement. Several superior high oleic peanut lines have been developed by marker assisted backcrossing in our breeding program. The objective of present study is to characterize bacterial wilt disease resistance and yield related traits in these novel peanut lines. The two-year disease nursery survival rates were used as an indicator for bacterial wilt resistance, and the diagnostic markers identified by QTL mapping were used for molecular verification. Genetic background of high oleic peanut lines was estimated by 40 K genotyping by target sequencing (GBTS) SNP panel. Preliminary and regional yield tests were performed for selected breeding lines in 2021. As a result, an elite high oleic peanut accession Yuhua183 was identified, inheriting the superior characteristics of its recurrent parent such as resistance to bacterial wilt, high shelling percentage and early maturity. The development of Yuhua183 broadened the genetic background of high oleic peanut germplasm, providing valuable genetic resources for peanut breeding and further utilization.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Peanut (Arachis hypogaea L.) is a valuable source of plant protein and edible oil worldwide. It is grown in more than 100 countries, with India and China ranking first in acreage and production, respectively (FAOSTAT 2020 https://www.fao.org/faostat/en/#home). Peanut oil is composed of eight fatty acids, namely palmitic acid (C16:0), stearic acid (C18:0), oleic acid (C18:1), linoleic acid (C18:2), behenic acid (C22:0), arachidic acid (C20:0), gadoleic acid (C20:1) and lignoceric acid (C24:0), of which oleic acid and linoleic acid account for approximately 80% of the total fatty acids (Shasidhar el al. 2017). The ratio of oleic acid and linoleic acid determines the quality of peanut oil. As unsaturated fatty acids, both oleic and linoleic acids are capable of lowering cholesterols and the risks of cardiovascular diseases. Linoleic acid encompasses one more reactive double bond than oleic acid, and is hence more prone to oxidation compared to oleic acid. Therefore, high oleic peanut oil is preferred in food industry because of chemical stability and long shelf life (Talcott et al. 2005a, b).
The direct conversion of oleic acid to linoleic acid was catalyzed by omega-6 fatty acid desaturase (namely FAD2) in seed oil of peanut (Ray et al. 1993; Lopez et al. 2001). Typically, the amount of oleic acid in peanut kernels ranges from 48 to 54%. A peanut mutant line F435, with the amount of oleic acid reaching 80%, was reported in 1987 (Norden et al. 1987). Mutations in FAD2, including single nucleotide substitution on chromosome A09 and nucleotide insertion on chromosome B09, result in marked attenuation in FAD2 activity and increased proportion of oleic acid at the expense of linoleic acid in F435 seed oil (Jung et al. 2000a, b). Consequently, a series of high oleic peanut cultivars were developed by using F435 as high oleic donor parent.
In the three decades of high oleic breeding in peanut, desirable traits such as disease resistance have been integrated in high oleic peanuts by marker assisted selection (Holbrook et al. 2017; Deshmukh et al. 2020). To facilitate the broadening of genetic background in high oleic peanut, we developed 34 advanced high oleic breeding lines by FAD2 marker assisted backcrossing (MABC) using high yielding, disease resistant and early matured elite peanut cultivars as recurrent parents (Huang et al. 2019). In the present study, we aim to characterize such elite lines derived from MABC and explore their utilization in high oleic peanut germplasm.
Materials and methods
Plant materials
Thirty-four high oleic peanut breeding lines developed by MABC, were selected to identify bacterial wilt resistance in a disease nursery (Table 1). Their recurrent parent Yuanza9102, is a Spanish-type peanut variety with bacterial wilt disease resistance released by the Institute of Industrial Crops, Henan Academy of Agricultural Sciences in 2001. A high oleic germplasm DF12, was the donor parent for high oleic alleles which were selected by fad2 markers in MABC (Huang et al. 2019).
Phenotyping for bacterial wilt resistance
High oleic breeding lines derived from Yuanza9102 were planted in a randomized complete block design with two replications in a bacterial wilt disease nursery (at Hezhou, Guangxi Province) in autumn 2018 and spring 2019. Twenty seeds of each accession were planted in row of 200 cm long with rows 30 cm apart. The numbers of emerging seedlings were recorded two weeks after sowing. At maturation, the established plants of each accession were counted and survival rates were calculated as an indicator of bacterial wilt resistance. Analysis of Variance (ANOVA) tests for survival rates of thirty-eight varieties were conducted by GenStat software 18th edition (VSN International Ltd.).
DNA extraction and marker genotyping
Genomic DNA was extracted from thin slices of mature seeds using the Plant Genomic DNA Kit (TIANGEN BIOTECH CO., Beijing, China). DNA concentrations were measured using NanoDrop 2000 (Thermo Scientific). The Kompetitive Allele Specific PCR (KASP) reaction mixture entailed 2.5 μL DNA template (10 to 20 ng), 2.43 μL of 2 × KASP Master Mix (LGC, Hoddesdon, UK), and 0.07 μL KASP Assay mix. The assay mix contains two allele-specific forward primers and one reverse primer (Table 2). Five breeding lines derived from Yuanza9102 were genotyped by KASP assay to identify bacterial wilt resistance using qBWA12 markers previously developed (Qi et al. 2022). All breeding lines derived from MABC were selected by fad2 KASP markers previously described (Huang et al. 2019).
Yield and quality traits of eight lines derived from Yuanza9102
Eight breeding lines were included in a preliminary test in the spring of 2021 at Yuanyang, Henan Province and planted in randomized complete block design with two replications. 160 seeds of each breeding line were sowed in plot consisted of four rows 40 cm apart and 600 cm long with plant space of 15 cm. Oleic acid content in the kernels of each breeding line was measured by a DA7250 near infrared analyzer (Perten Instruments, Sweden), as previously described (Huang et al. 2019). Yield traits including hundred-pod weight (HPW), hundred-kernel weight (HKW) and shelling percentage were investigated. One-way Analysis of Variance (ANOVA) tests for yield data of eight breeding lines were conducted by GenStat software 18th edition (VSN International Ltd.).
Phylogenetic analysis of five Yuanza9102 derived lines
Eleven accessions including five Yuanza9102 derived lines were genotyped by 40 K SNP panel developed from genotyping by target sequencing (GBTS). A phylogenetic tree was constructed by Neighbor-Joining method with 1000 bootstrap replications in MEGA software (Kumar et al. 2018). The tree was drawn by online tool iTOL v6 (https://itol.embl.de/).
Multi-location test
Among the high oleic lines derived from Yuanza9102, HAAS2BC4-140(N)-11–2–3, also known as Yuhua183, was included in the yield test in Jiangxi Province with Changhua1 (CH1) as the check cultivar. Eight accessions were planted in a randomized complete block design with three replications at five locations in 2021. Each plot was 5 m long and 2 m wide. 300 seeds of each accession were sowed per plot. Ten plants from each plot were collected for phenotyping. Maturity date was recorded for each accession. Yield traits were estimated by HPW, HKW and shelling percentage. The whole plots were harvested for yield evaluation. Analysis of Variance (ANOVA) tests for yield data of eight varieties were conducted by GenStat software 18th edition (VSN International Ltd.).
Results
Bacterial wilt resistance of Yuanza9102 derived lines
In spring 2018 and autumn 2019, 34 breeding lines derived from marker assisted backcrossing were tested in the bacterial wilt disease nursery at Hezhou, Guangxi Province (Table 1). Survival rates of the breeding lines were not different from those of recurrent parent Yuanza9102 and resistant check Zhonghua6. These derived lines were distinct from susceptible check Zhonghua12 and high oleic donor parent DF12 based on survival rates (Supplementary Table S1). However, significant variation of survival rates was observed between the years of 2018 and 2019 (Supplementary Table S2).
Variation of yield and quality traits
Eight superior lines derived from Yuanza9102 were investigated for yield related traits in a preliminary test at Yuanyang, Henan province in 2021 (Table 1). The shelling percentage of these lines were not significantly different from one another (Table 3; Supplementary Table S3). Significant variations of HPW and HKW were observed among eight breeding lines. The HPW of HAAS2BC4-140(N)-11–2–3 was significantly larger than that of Yuanza9102. The HKW of two breeding lines were significantly lower than that of their recurrent parent, while the HKW of other six lines were similar to Yuanza9102. The oleic acid content of eight Yuanza9102 derived lines was between 72.52 and 75.23%, significantly higher than that of Yuanza9102.
Genetic characterization of Yuanza9102 derived lines
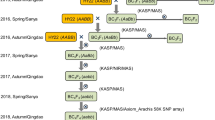
Five superior lines derived from Yuanza9102 were selected and genotyped by 40 K SNP panel (Table 1). Three lines derived from Yuhua15 were also included. The 40 K SNP data of eleven accessions was filtered by minor allele frequency (MAF) of 0.01, resulting in 11,435 SNPs. Phylogenetic analysis showed that these breeding lines were clearly grouped in association with their respective recurrent parent. HAAS2BC4-140(N)-4–10–3 and Yuanza9102 formed a single clade, indicating their close genetic association with each other, distinct from other Yuanza9102 derived lines. HAAS2BC4-140(N)-11–2–3 and HAAS2BC4-118(N)-2–1–1 were both found on separate branches distinct from Yuanza9102, with HAAS2BC4-140(N)-11–2–3 clustered more closely with Yuanza9102 (Fig. 1).
The five lines derived from Yuanza9102 were examined by SNP markers closely linked to bacterial wilt resistance. Their genotypes were the same as the genotype of bacterial wilt resistant parent Yuanza9102 based on KASP results at five SNP loci (Table 4). The five SNPs of high oleic donor parent DF12 were different from those of bacterial wilt resistant accessions, indicating that alleles from recurrent parent Yuanza9102 contributed to bacterial wilt resistance in these derived lines.
Yield related traits of Yuhua183 in multi-location test
Yuhua183, namely the breeding line HAAS2BC4-140(N)-11–2–3, matured 4 days earlier than the check cultivar CH1 in the multi-location test in Jiangxi province in 2021 (Table 5). The average shelling percentage of Yuhua183 reached 71.35%, which was the highest among eight varieties (Supplementary Tables S4 and S5). The pod yield and kernel yield (kg/ha) of Yuhua183 were not significantly different from those of the check cultivar at five locations.
Discussion
A spontaneous peanut mutant F435 with elevated oleic acid content was discovered in the U.S. breeding program (Norden et al. 1987). Since then, F435 and its derived lines had been used as the high oleic donor lines in the U.S., leading to the release of several high oleic peanut varieties including ‘SunOleic 95R’, ‘Tamrun OL01’, ‘Tamrun OL02’, ‘Tamrun OL12’ and ‘Florida-07’ (Burow et al. 2014; Gorbet and Knauft 1997; Gorbet and Tillman 2009; Simpson et al. 2003, 2006). The early-maturing variety “Tamrun OL12”, was derived from a single cross between a high oleic runner-type breeding line and an early maturing Spanish-type landrace. In more recent breeding schemes for high oleic peanuts, high oleic trait was pyramided with disease resistance or high oil content by marker assisted selection and marker assisted backcrossing (Chu et al. 2011; Janila et al. 2016; Holbrook et al. 2017; Huang et al. 2019).
Runner-type high oleic peanut accessions released in the U.S. are usually with long growth duration and spreading growth habit. The two-crop rotation cultivation in China is not suitable for planting runner-type peanuts. Cultivars released in China required shorter growth duration and are usually with erect or bunch growth habit. The goal of peanut breeding in China is high yield with growth duration around 120 days adapted to the two-crop rotation system. When the U.S. high oleic peanut germplasm were crossed with Chinese peanut cultivars, the progenies would show wide segregation of yield related traits, such as growth habit, growth duration, and seed size. Based on a survey of nine released high oleic varieties in the first decades (from year of 2007 to 2017) since introduction of high oleic peanut germplasm, the growth duration of five varieties were longer than 125 days, while the HKW of six varieties were less than 80 g (Wang et al. 2021). Moreover, it is imperative to pyramid disease resistance with high oleic trait to adapt to field conditions with disease and pests. Marker assisted backcrossing is an effective approach to increase oleic acid content in locally adapted cultivars. In the backcross breeding program, our goal is to increase oleic acid content of popular peanut varieties in China. The recurrent parents are usually with erect branching habit and matured earlier than the high oleic donor parents.
To develop high oleic peanut varieties with bacterial wilt resistance, Yuanza9102 was chosen as the recurrent parent in our marker assisted backcross breeding project. Among the advanced lines derived from Yuanza9102, Yuhua183 was selected due to its high oleic acid content and early maturity. Yuhua183 and its recurrent parent Yuanza9102 share competitive characteristics including high shelling percentage. In the multi-location test, the pod yield and kernel yield of Yuhua183 were not significantly different from those of the check cultivar. Genotyping results of 40 K SNP panel and bacterial wilt diagnostic KASP markers indicated that Yuhua183 is a high oleic peanut accession genetically similar to Yuanza9102 and is identified as resistant to bacterial wilt disease.
Conclusion
An elite high oleic peanut cultivar, Yuhua183, was developed by FAD2 marker assisted backcrossing using popular cultivar Yuanza9102 as recurrent parent. The bacterial wilt resistance was evaluated in disease nursery for two seasons, which was validated by qBWA12 diagnostic SNP markers. Phenotypic data at five locations indicated that Yuhua183 matures early with stable yield and high shelling percentage. Yuhua183 is an elite peanut cultivar which expands the genetic background of high oleic peanuts, and is a valuable germplasm resource for further utilization in peanut breeding.
References
Burow MD, Baring MR, Ayers JL, Schubert AM, Lopez Y, Simpson CE (2014) Registration of ‘Tamrun OL12’ peanut. J Plant Regist 8:117
Chu Y, Wu CL, Holbrook CC, Tillman BL, Person G, Ozias-Akins P (2011) Marker assisted selection to pyramid nematode resistance and the high Oleic trait in Peanut. Plant Genome 4:110–117
Deshmukh DB, Marathi B, Sudini HK, Variath MT, Chaudhari S, Manohar SS, Rani CVD, Pandey MK, Pasupuleti J (2020) Combining high oleic acid trait and resistance to late leaf spot and rust diseases in groundnut (Arachis hypogaea L.). Front Genet 11:514
Gorbet DW, Knauft DA (1997) Registration of “SunOleic 95R” peanut. Crop Sci 37:1392
Gorbet DW, Tillman BL (2009) Registration of ‘Florida-07 peanut. J Plant Regist 3:14–18
Holbrook CC, Ozias-Akins P, Chu Y, Culbreath AK, Kvien CK, Brenneman TB (2017) Registration of ‘TifNV-High O/L’ Peanut. J Plant Regist 11:228–230
Huang B, Qi F, Sun Z, Miao L, Zhang Z, Liu H, Fang Y, Dong W, Tang F, Zheng Z, Zhang X (2019) Marker-assisted backcrossing to improve seed oleic acid content in four elite and popular peanut (Arachis hypogaea L.) cultivars with high oil content. Breed Sci 69:234–243
Janila P, Pandey MK, Shasidhar Y, Variath MT, Sriswathi M, Khera P, Manohar SS, Nagesh P, Vishwakarma MK, Mishra GP, Radhakrishnan T, Manivannan N, Dobariya KL, Vasanthi RP, Varshney RK (2016) Molecular breeding for introgression of fatty acid desaturase mutant alleles (ahFAD2A and ahFAD2B) enhances oil quality in high and low oil containing peanut genotypes. Plant Sci 242:203–213
Jung S, Powell G, Moore K, Abbott A (2000a) The high oleate trait in the cultivated peanut (Arachis hypogaea L.): II. Molecular basis and genetics of the trait. Mol Gen Genet 263:806–811
Jung S, Swift D, Sengoku E, Patel M, Teule F, Powell G, Moore K, Abbott A (2000b) The high oleate trait in the cultivated peanut (Arachis hypogaea L.): I. Isolation and characterization of two genes encoding microsomal oleoyl-PC desaturases. Mol Gen Genet 263:796–805
Kumar S, Stecher G, Li M, Knyaz C, Tamura K (2018) MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol 35:1547–1549
Lopez Y, Smith OD, Senseman SA, Rooney WL (2001) Genetic factors influencing high oleic acid content in Spanish market-type peanut cultivars. Crop Sci 41:51–56
Norden AJ, Gorbet DW, Knauft DA, Young CT (1987) Variability in oil quality among peanut genotypes in the Florida breeding program. Peanut Sci 14:7–11
Qi F, Sun Z, Liu H, Zheng Z, Qin L, Shi L, Chen Q, Liu H, Lin X, Miao L, Tian M, Wang X, Huang B, Dong W, Zhang X (2022) QTL identification, fine mapping, and marker development for breeding peanut (Arachis hypogaea L.) resistant to bacterial wilt. Theor Appl Genet. https://doi.org/10.1007/s00122-022-04033-y
Ray TK, Holly SP, Knauft DA, Abbott AG, Powell GL (1993) The primary defect in developing seed from the high oleate variety of peanut (Arachis hypogaea L.) is the absence of Δ12-desaturase activity. Plant Sci 91:15–21
Shasidhar Y, Vishwakarma MK, Pandey MK, Janila P, Variath MT, Manohar SS, Nigam SN, Guo B, Varshney RK (2017) Molecular mapping of oil content and fatty acids using dense genetic maps in groundnut (Arachis hypogaea L.). Front Plant Sci 8:794
Simpson CE, Baring MR, Schubert AM, Melouk HA, Black MC, Lopez Y, Keim KA (2003) Registration of ‘Tamrun OL01’ peanut. Crop Sci 43:2298
Simpson CE, Baring MR, Schubert AM, Black MC, Melouk HA, Lopez Y (2006) Registration of “Tamrun OL02” peanut. Crop Sci 46:1813
Talcott ST, Duncan CE, Pozo-Insfran DD, Gorbet DW (2005a) Polyphenolic and antioxidant changes during storage of normal, mid, and high oleic acid peanuts. Food Chem 89:77–84
Talcott ST, Passeretti S, Duncan CE, Gorbet DW (2005b) Polyphenolic content and sensory properties of normal and high oleic acid peanuts. Food Chem 90:379–388
Wang C, Yu S, Zhu L (2021) High oleic acid peanuts in China. Shanghai Science and Technology Press, Shanghai
Funding
This work was supported by China Agriculture Research System (CARS-13), Henan Provincial Agriculture Research System, China (S2012-5), Major Science and Technology Projects of Henan Province (201300111000), and the Henan Provincial R&D Projects of Interregional Cooperation for Local Scientific and Technological Development Guided by Central Government (YDZX20214100004191).
Author information
Authors and Affiliations
Contributions
XZ and BH were responsible for the research design and supervising. YF drafted the manuscript and analyzed the data. Field work were conducted by HL, QC, HL, FQ, ZZ, BH, LW, WD. Lab work was conducted by XW, MT, FQ, DL, ZS. All authors approved the final manuscript.
Corresponding authors
Ethics declarations
Conflict of interest
Authors declare no conflict of interests and the manuscript is approved by all authors for publication.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Fang, Y., Liu, H., Zhang, Z. et al. Genomic identification and phenotypic evaluation of yield traits and bacterial wilt resistance in high oleic peanut breeding lines developed by marker assisted backcrossing. Euphytica 218, 80 (2022). https://doi.org/10.1007/s10681-022-03032-w
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s10681-022-03032-w