Abstract
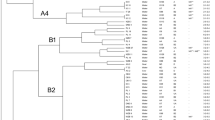
Genotypic diversity among multi-drug-resistant (MDR) aquatic E. coli isolated from different sites of Yamuna River was analyzed using repetitive element PCR (rep-PCR) methods viz. ERIC-PCR and (GTG)5-PCR and compared with the MDR animal fecal isolates. The 97 E. coli isolates belonging to different serotypes, phylogroups, and multi-drug resistance patterns were analyzed. High genetic diversity was observed by both the methods; however, (GTG)5 typing showed higher discriminating potential. Combination of ERIC types (E1–E32) and (GTG)5 types (G1–G46) generated 77 genotypes. The frequency of genotypes ranged from 0.013 to 0.065. The genotype composition of E. coli isolates was highly diverse at all the sampling sites across Yamuna River except at its entry site in Delhi. The sampling sites under the influence of high anthropogenic activities showed an increase in number of unique genotype isolates. These sites also exhibited high multiple antibiotic resistance (MAR) indexes (above 0.25) suggesting high risk of contamination. Principal coordinate analysis (PCoA) showed limited clustering of genotypes based on the sampling sites. The most frequent genotypes were grouped in the positive zone of both the principal coordinates (PC1 and PC2). The genotypes of most of the animal fecal isolates were unique and occupied a common space in the negative PC1 area forming a separate cluster. High genotypic diversity among the aquatic E. coli and the drain isolates, discharging the untreated municipal waste in the river, was observed, suggesting that the sewage effluents contribute substantially to contamination of this river system than animal feces. The presence of such a high diversity among the MDR E. coli isolates in the natural river systems is of great public health significance and highlights the need of an efficient surveillance system for better management of Indian natural water bodies.



Similar content being viewed by others
References
Aristizábal-Hoyos, A. M., Rodríguez, E. A., Arias, L., & Jiménez, J. N. (2019). High clonal diversity of multidrug-resistant and extended spectrum beta-lactamase-producing Escherichia coli in a wastewater treatment plant. Journal of Environmental Management, 245, 37–47.
Azam, M., Jan, A. T., Kumar, A., Siddiqui, K., Mondal, A. H., & Haq, Q. M. R. (2018). Study of pandrug and heavy metal resistance among E. coli from anthropogenically influenced Delhi stretch of river Yamuna. Brazilian Journal of Microbiology, 49(3), 471–480.
Baldy-Chudzik, K., & Stosik, M. (2005). Specific genomic fingerprints of Escherichia coli strains with repetitive sequences and PCR as an effective tool for monitoring freshwater environments. Polish Journal of Environmental Studies, 14, 551–557.
Bergholz, P. W., Noar, J. D., & Buckley, D. H. (2011). Environmental patterns are imposed on the population structure of Escherichia coli after fecal deposition. Applied and Environmental Microbiology, 77(1), 211–219.
Beyer, W., Mukendi, F. M., Kimmig, P., & Bohm, R. (1998). Suitability of repetitive-DNA-sequence-based PCR fingerprinting for characterizing epidemic isolates of Salmonella enterica serovar Saintpaul. Journal of Clinical Microbiology, 36(6), 1549–1554.
Bidhuri, S., & Jain, P. (2019). Identifying waterborne disease prone areas using geospatial approach along the right bank of Yamuna River in Delhi. International Journal of Environmental Health Research, 29(5), 561–581.
Bogaerts, P., Rodriguez-Villalobos, H., Bauraing, C., Deplano, A., Laurent, C., Berhin, C., Struelens, M. J., & Glupczynski, Y. (2010). Molecular characterization of AmpC-producing Escherichia coli clinical isolates recovered at two Belgian hospitals. Pathologie Biologie, 58(1), 78–83.
Borges, L. G. D. A., Dalla Vechia, V., & Corçóo, G. (2003). Characterisation and genetic diversity via REP-PCR of Escherichia coli isolates from polluted waters in southern Brazil. FEMS Microbiology Ecology, 45(2), 173–180.
Cook, K. L., Bolster, C. H., Ayers, K. A., & Reynolds, D. N. (2011). Escherichia coli diversity in livestock manures and agriculturally impacted stream waters. Current Microbiology, 63(5), 439–449. https://doi.org/10.1007/s00284-011-0002-6.
De Vuyst, L., Camu, N., De Winter, T., Vandemeulebroecke, K., Van de Perre, V., Vancanneyt, M., et al. (2008). Validation of the (GTG)5-rep-PCR fingerprinting technique for rapid classification and identification of acetic acid bacteria, with a focus on isolates from Ghanaian fermented cocoa beans. International Journal of Food Microbiology, 125(1), 79–90.
Dubey, R. S. (2016). Assessment of water quality status of Yamuna river and its treatment by electrode based techniques. International Journal of Engineering Sciences & Research Technology, 5(2), 448–455.
Fogarty, L. R., Haack, S. K., Wolcott, M. J., & Whitman, R. L. (2003). Abundance and characteristics of the recreational water quality indicator bacteria Escherichia coli and enterococci in gull faeces. Journal of Applied Microbiology, 94(5), 865–878.
Gevers, D., Huys, G., & Swings, J. (2001). Applicability of rep-PCR fingerprinting for identification of Lactobacillus species. FEMS Microbiology Letters, 205(1), 31–36.
Goto, D. K., & Yan, T. (2011). Genotypic diversity of Escherichia coli in the water and soil of tropical watersheds in Hawaii. Applied And Environmental Microbiology, 77(12), 3988–3997.
Grundmann, H., Hori, S., & Tanner, G. (2001). Determining confidence intervals when measuring genetic diversity and the discriminatory abilities of typing methods for microorganisms. Journal of Clinical Microbiology, 39(11), 4190–4192.
Haberman, D. L. (2006). River of love in an age of pollution: the Yamuna River of northern India. Berkeley: University of California Press.
Hashemi, A., & Baghbani-arani, F. (2015). The effective differentiation of Salmonella isolates using four PCR-based typing methods. Journal of Applied Microbiology, 118(6), 1530–1540.
Hu, Y., Cai, J., Zhou, H., Chi, D., Zhang, X., Chen, W., et al. (2013). Molecular typing of CTX-M-producing Escherichia coli isolates from environmental water, swine feces, specimens from healthy humans, and human patients. Applied and Environmental Microbiology, 79(19), 5988–5996.
Hunter, P. R., & Gaston, M. A. (1988). Numerical index of the discriminatory ability of typing systems: an application of Simpson’s index of diversity. Journal of Clinical Microbiology, 26(11), 2465–2466.
Iacumin, L., Comi, G., Cantoni, C., & Cocolin, L. (2006). Molecular and technological characterization of Staphylococcus xylosus isolated from naturally fermented Italian sausages by RAPD, Rep-PCR and Sau-PCR analysis. Meat Science, 74(2), 281–288.
Kaushik, M., Khare, N., Kumar, S., & Gulati, P. (2019). High prevalence of antibiotic resistance and integrons in Escherichia coli isolated from urban river water, India. Microbial Drug Resistance, 25(3), 359–370.
Khare, N., Kaushik, M., Kumar, S., & Gulati, P. (2020). Evaluation of genetic diversity among aquatic and fecal isolates of Escherichia coli using multilocus variable number of tandem repeat analysis. 3 Biotech, 10(2), 63.
Kon, T., Weir, S. C., Howell, E. T., Lee, H., & Trevors, J. T. (2009). Repetitive element (REP)-polymerase chain reaction (PCR) analysis of Escherichia coli isolates from recreational waters of southeastern Lake Huron. Canandian Journal of Microbiology, 55(3), 269–276.
Krumperman, P. (1983). Multiple antibiotic resistance indexing of Escherichia coli to identify high-risk sources of fecal contamination of foods. Applied and Environmental Microbiology, 46, 165–170.
Labrador, K. L., Nacario, M. A. G., Malajacan, G. T., Abello, J. J. M., Galarion, L. H., Rensing, C., & Rivera, W. L. (2020). Selecting rep-PCR markers to source track fecal contamination in Laguna Lake, Philippines. Journal of Water and Health, 18(1), 19–29.
Lan, R., & Reeves, P. R. (2000). Intraspecies variation in bacterial genomes: the need for a species genome concept. Trends in Microbiology, 8(9), 396–401.
Londero, A., Costa, M., Sucari, A., & Leotta, G. (2019). Comparison of three molecular subtyping techniques for Listeria monocytogenes. Revista Argentina de Microbiologia, 51(4), 359–362.
Lyautey, E., Lu, Z., Lapen, D. R., Berkers, T. E., Edge, T. A., & Topp, E. (2010). Optimization and validation of rep-PCR genotypic libraries for microbial source tracking of environmental Escherichia coli isolates. Canadian Journal of Microbiology, 56(1), 8–17.
Ma, H., Fu, L., & Li, J. (2011). Differentiation of fecal Escherichia coli from human, livestock, and poultry sources by rep-PCR DNA fingerprinting on the shellfish culture area of East China Sea. Current Microbiology, 62(5), 1423–1430.
McLellan, S. L., Daniels, A. D., & Salmore, A. K. (2003). Genetic characterization of Escherichia coli populations from host sources of fecal pollution by using DNA fingerprinting. Applied and Environmental Microbiology, 69(5), 2587–2594.
Mohapatra, B. R., & Mazumder, A. (2008). Comparative efficacy of five different rep-PCR methods to discriminate Escherichia coli populations in aquatic environments. Water Science and Technology, 58(3), 537–547.
Mohapatra, B. R., Broersma, K., & Mazumder, A. (2007). Comparison of five rep-PCR genomic fingerprinting methods for differentiation of fecal Escherichia coli from humans, poultry and wild birds. FEMS Microbiology Letters, 277(1), 98–106.
Murugan, K., Prabhakaran, P., Al-Sohaibani, S., & Sekar, K. (2012). Identification of source of faecal pollution of Tirumanimuttar River, Tamilnadu, India using microbial source tracking. Environmental Monitoring and Assessment, 184(10), 6001–6012.
Nicholas, A., Kim, Y. K., Lee, W. K., Selasi, G. N., Na, S. H., Kwon, H. I., Kim, Y. J., Lee, H. S., Song, K. E., Shin, J. H., & Lee, J. C. (2017). Molecular epidemiology and antimicrobial susceptibility of Clostridium difficile isolates from two Korean hospitals. PLoS One, 12(3), e0174716.
Rohlf, F. J. (2008). NTSYSpc: Numerical Taxonomy System, ver, 2.20. Setauket: Exeter Publishing Ltd.
Sabat, A. J., Budimir, A., Nashev, D., Sá-Leão, R., van Dijl, J. M., Laurent, F., et al. (2013). Overview of molecular typing methods for outbreak detection and epidemiological surveillance. Eurosurveillance, 18(4), 1–15.
Sharma, D., & Kansal, A. (2011). Water quality analysis of River Yamuna using water quality index in the national capital territory, India (2000–2009). Applied Water Science, 1, 147–157.
Sprenger, C., Lorenzen, G., Grunert, A., Ronghang, M., Dizer, H., Selinka, H. C., Girones, R., Lopez-Pila, J. M., Mittal, A. K., & Szewzyk, R. (2014). Removal of indigenous coliphages and enteric viruses during riverbank filtration from highly polluted river water in Delhi (India). Journal of Water and Health, 12(2), 332–342.
Švec, P., Vancanneyt, M., Seman, M., Snauwaert, C., Lefebvre, K., Sedláček, I., et al. (2005). Evaluation of (GTG)5-PCR for identification of Enterococcus spp. FEMS Microbiology Letters, 247(1), 59–63.
Tikoo, A., Tripathi, A. K., Verma, S. C., Agrawal, N., & Nath, G. (2001). Application of PCR fingerprinting techniques for identification and discrimination of Salmonella isolates. Current Science, 80(8), 1049–1052.
Versalovic, J., Koeuth, T., & Lupski, J. R. (1991). Distribution of repetitive DNA sequences in eubacteria and application to fingerprinting of bacterial genomes. Nucleic Acids Research, 19(24), 6823–6831.
Versalovic, J., Schneider, M., de Bruijn, F. J., & Lupski, J. R. (1994). Genomic fingerprinting of bacteria using repetitive sequence-based polymerase chain reaction. Methods in Molecular and Cellular Biology, 5, 25–40.
Weir, B. S. (1990). Genetic data analysis: methods for discrete population genetic data analysis. Sunderland: Sinauer Associates Inc Publishers.
Acknowledgments
Financial assistance received from the University Grant Commission (UGC) (Major Research Project grant no. UGC-41-1172/2012) and the Department of Science and Technology (DST grant no. 1196 SR/FST/LS-I/2017/4) is duly acknowledged. NK and MK are grateful to Maharshi Dayanand University for providing University Research Scholarship. NK sincerely thanks UGC for RGNF grant (UGC-RGNF grant no. F1-17.1/2016-17/RGNF-2015-17-SC-HAR-18413/(SA-III/Website)) and MK thanks CSIR for the Senior Research Fellowship (no. 09/382(0212)/19-EMR-I). The authors also wish to acknowledge National Salmonella and Escherichia Centre, Central Research Institute, Kasauli, Himachal Pradesh, India, for serotyping E. coli isolates.
Funding
This study was funded by the University Grant Commission (UGC) (Major Research Project grant no. UGC-41-1172/2012) and the Department of Science and Technology (grant no. 1196 SR/FST/LS-I/2017/4).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethics approval
Not applicable.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
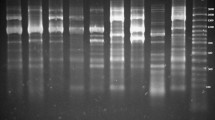
Supplementary Fig 1.
Representative gels showing (A) ERIC- fingerprints (Lane 1-17) and (B) (GTG)5- fingerprints (Lane 18-34) of E. coli obtained in the present study. (JPG 968 kb)
Rights and permissions
About this article
Cite this article
Khare, N., Kaushik, M., Martin, J.P. et al. Genotypic diversity in multi-drug-resistant E. coli isolated from animal feces and Yamuna River water, India, using rep-PCR fingerprinting. Environ Monit Assess 192, 681 (2020). https://doi.org/10.1007/s10661-020-08635-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s10661-020-08635-1