Abstract
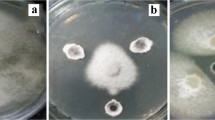
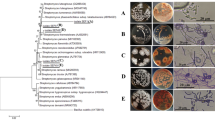
Eighty-six isolates of Streptomyces-like colonies were isolated from soil, and their antagonistic activity was assessed against Fusarium oxysporum f. sp. lycopersici (Fol) as well as their plant growth-promoting (PGP) activity. The results showed that five isolates could inhibit the mycelial growth of the fungal pathogen by producing extracellular hydrolytic enzymes for cellulose and amylase. The enzyme activities cause rough cell walls, irregular swelling, membrane collapse, and nuclear membrane and chromatin fiber degradation. In the pot experiment, the culture filtrates and cell suspensions from all isolates could inhibit fungal pathogens in tomato plants. Among them, isolate S.PNR29 had the best ability to protect the tomato plants from fungal pathogens with low disease severity scores, lowest disease severity index, and highest disease control efficacy. Moreover, these isolates had IAA production, phosphate solubilization, and siderophore production to enhance the tomato growth under pot experiments, especially isolate S.PNR29, which enhanced tomato growth and seed germination rate. These potential antagonistic strains were preliminarily identified using 16S rRNA gene sequencing. The 16S rRNA gene sequences confirmed these strains as bacteria of the genus Streptomyces. The best antagonistic and plant-growth-promoting isolate S.PNR29 was identified by multi-locus sequence analysis (MLSA) and whole genome sequencing. Interestingly, biosynthetic gene clusters (BGCs) related to antibiotics and gene clusters related to plant-growth-promoting and antagonistic properties were found in the genome of Streptomyces sp. isolate S.PNR29. These data may be valuable and used to support the antagonistic activity of Streptomyces sp. isolate S.PNR29 against the Fusarium wilt pathogen and PGP abilities in tomatoes.






Similar content being viewed by others
Data Availability
The data supporting this study's findings are available from the corresponding author upon reasonable request.
References
Al-Askar, A. A., Baka, Z. A., Rashad, Y. M., Ghoneem, K. M., Abdulkhair, W. M., Hafez, E. E., & Shabana, Y. M. (2015). Evaluation of Streptomyces griseorubens E44G for the biocontrol of Fusarium oxysporum f. sp. lycopersici: ultrastructural and cytochemical investigations. Annals of Microbiology, 65(4), 1815–1824. https://doi.org/10.1007/s13213-014-1019-4
Anitha, A., & Rabeeth, M. (2009). Control of Fusarium wilt of tomato by bioformulation of Streptomyces griseus in green house condition. African Journal of Basic & Applied Sciences, 1(1–2), 9–14.
Anusha, B. G., Gopalakrishnan, S., Naik, M. K., & Sharma, M. (2019). Evaluation of Streptomyces spp. and Bacillus spp. for biocontrol of Fusarium wilt in chickpea (Cicer arietinum L.). Archives of Phytopathology and Plant Protection, 52(5–6), 417–442. https://doi.org/10.1080/03235408.2019.1635302
Branthôme, F.X. (2022) Worldwide (Total Fresh) Tomato Production Exceeds 187 Million Tonnes in 2020. Available online: https://www.tomatonews.com/en/worldwide-total-fresh-tomato-production-exceeds-187-million-tonnes-in-2020_2_1565.html (accessed on 12 April 2023)
Bubici, G. (2018). Streptomyces spp. as biocontrol agents against Fusarium species. CABI Reviews, 2018, 1–15. https://doi.org/10.1079/PAVSNNR20181305
Chaiharn, M., Theantana, T., & Pathom-Aree, W. (2020). Evaluation of biocontrol activities of Streptomyces spp. against rice blast disease fungi. Pathogens, 9(2), 126. https://doi.org/10.3390/pathogens9020126
Chakraborty, M., Mahmud, N. U., Md. Muzahid, A. N., Fajle Rabby, S. M., & Islam, T. (2020). Oligomycins inhibit Magnaporthe oryzae Triticum and suppress wheat blast disease. PlosONE. https://doi.org/10.1371/journal.pone.0233665
Chun, J., Oren, A., Ventosa, A., Christensen, H., Arahal, D. R., da Costa, M. S., Rooney, A. P., Yi, H., Xu, X. W., De Meyer, S., & Trujillo, M. E. (2018). Proposed minimal standards for the use of genome data for the taxonomy of prokaryotes. International Journal of Systematic and Evolutionary, 68(1), 461–466. https://doi.org/10.1099/ijsem.0.002516
Codd, R., Richardson-Sanchez, T., Telfer, T. J., & Gotsbacher, M. P. (2018). Advances in the chemical biology of desferrioxamine B. ACS Chemical Biology, 13(1), 11–25. https://doi.org/10.1021/acschembio.7b00851
Conn, V. M., Walker, A. R., & Franco, C. M. M. (2008). Endophytic actinobacteria induce defense pathways in Arabidopsis thaliana. Molecular Plant-Microbe Interactions, 21(2), 208–218. https://doi.org/10.1094/MPMI-21-2-0208
Das, I. K., Indira, S., Annapurna, A., & Seetharama, N. (2008). Biocontrol of charcoal rot in sorghum by fluorescent Pseudomonads associated with the rhizosphere. Crop Protection, 27(11), 1407–1414. https://doi.org/10.1016/j.cropro.2008.07.001
Domsch, K. H., Jagnow, G., & Anderson, T. H. (1983). An ecological concept for the assessment of side-effects of agrochemicals on soil microorganisms. Residue Reviews., 86, 65–105.
Erdogan, O., & Benlioglu, K. (2010). Biological control of Verticillium wilt on cotton by the use of Fluorescent Pseudomonas sp. under field conditions. Biological Control, 53(1), 39–45. https://doi.org/10.1016/j.biocontrol.2009.11.011
Glickmann, E., & Dessaux, Y. (1995). A critical examination of the specificity of the Salkowski reagent for indolic compounds produced by phytopathogenic bacteria. Applied and Environmental Microbiology, 61(2), 793–796. https://doi.org/10.1128/aem.61.2.793-796.1995
Gu, S., Yang, T., Shao, Z., Wang, T., & Pommier, T. (2020). Siderophore-mediated interactions determine the disease suppressiveness of microbial consortia. mSystems, 5, 811–819. https://doi.org/10.1128/mSystems.00811-19
Hao, D., Gao, P., Liu, P., Zhao, J., Wang, Y., Yang, W., Lu, Y., Shi, T., & Zhang, X. (2011). AC3-33 a novel secretory protein, inhibits Elk1 transcriptional activity via ERK pathway. Molecular Biology Reports, 38, 1375–1382.
Hibar, K., Daami-Remadi, M., Jabnoun-Khiareddine, H., & El Mahjoub, M. (2006). Temperature effect on mycelial growth and on disease incidence of Fusarium oxysporum f. sp. radicis-lycopersici. Plant pathology journal, 5(2), 233–238. https://doi.org/10.3923/ppj.2006.233.238
Kaewkla, O., Sukpanoa, S., Suriyachadkun, C., Chamroensaksi, N., Chumroenphat, T., & Franco, C. M. M. (2022). Streptomyces spinosus sp. nov. and Streptomyces shenzhenensis subsp. oryzicola subsp. nov. endophytic actinobacteria isolated from Jasmine rice and their genome mining for potential as antibiotic producers and plant growth promoters. Antonie Van Leeuwenhoek, 115, 871–888. https://doi.org/10.1007/s10482-022-01741-9
Kamal, R., & Sharma, A. K. (2014). Control of Fusarium wilt using biocontrol agent Streptomyces sp. CPP-53 isolated from compost with plant growth promoting effect on tomato under greenhouse condition. Journal of Microbiology and Antimicrobials, 6(6), 97–103.
Kämpfer, P. (2012). Genus I. Streptomyces Waksman & Henrici 1943, 339AL emend. Witt & Stackebrandt 1990, 370 emend. Wellington, Stackebrandt, Sanders, Wolstrup & Jorgensen 1992, 159 In: Whitman W. B., Goodfellow M., Kämpfer P., Busse H. J., Trujillo M. E., Ludwig W., Suzuki K. I., & Parte A. (eds.) Bergey’s Manual of Systematic Bacteriology, 2nd (edn): vol. 4, New York, Springer 1750 pp
Kawicha, P., Laopha, A., Chamnansing, W., Sopawed, W., Wongcharone, A., & Sangdee, A. (2020). Biocontrol and plant growth-promoting properties of Streptomyces isolated from vermicompost soil. Indian Phytopathology, 73(4), 655–666. https://doi.org/10.1007/s42360-020-00267-2
Kawicha, P., Nitayaros, J., Saman, P., Thaporn, S., Thanyasiriwat, T., Somtrakoon, K., Sangdee, K., & Sangdee, A. (2023a). Evaluation of Soil Streptomyces spp. for the Biological Control of Fusarium Wilt Disease and Growth Promotion in Tomato and Banana. The Plant Pathology Journal, 39(1), 108–122.
Kawicha, P., Tongyoo, P., Wongpakdee, S., Rattanapolsan, L., Duangjit, J., Chunwongse, J., Suwor, P., Sangdee, A., & Thanyasiriwat, T. (2023b). Genome-wide association study revealed genetic loci for resistance to fusarium wilt in tomato germplasm. Crop Breeding and Applied Biotechnology, 23, e43532311. https://doi.org/10.1590/1984-70332023v23n1a1
Kimura, M. (1980). A simple method for estimating evolutionary rate of base substitutions through comparative studies of nucleotide sequences. Journal of Molecular Evolution, 16(2), 111–120. https://doi.org/10.1007/BF01731581
Kumar, A. S., Reddy, N. E., Reddy, K. H., & Devi, M. C. (2007). Evaluation of fungicidal resistance among Colletotrichum gloeosporioides isolates causing mango anthracnose in Agri Export Zone of Andhra Pradesh. India. Plant Pathology Bulletin, 16(3), 157–160.
Law, J. W. F., Pusparajah, P., Ab Mutalib, N. S., Wong, S. H., Goh, B. H., & Lee, L. H. (2019). A review on mangrove actinobacterial diversity the roles of Streptomyces and novel species discovery. Progress In Microbes & Molecular Biology, 2(1), a0000024. https://doi.org/10.36877/pmmb.a0000024
Le, K. D., Yu, N. H., Park, A. R., Park, D. J., Kim, C. J., & Kim, J. C. (2022). Streptomyces sp. AN090126 as a biocontrol agent against bacterial and fungal plant diseases. Microorganisms, 10(4), 791. https://doi.org/10.3390/microorganisms10040791
Lefort, V., Desper, R., & Gascuel, O. (2015). FastME 2.0: a comprehensive, accurate, and fast distance-based phylogeny inference program. Molecular biology and evolution, 32(10), 2798–2800. https://doi.org/10.1093/molbev/msv150
Lynd, L. R., Weimer, P. J., Van Zyl, W. H., & Pretorius, I. S. (2002). Microbial cellulose utilization: Fundamentals and biotechnology. Microbiology and Molecular Biology Reviews, 66(3), 506–577. https://doi.org/10.1128/MMBR.66.3.506-577.2002
Marlatt, M. L., Correll, J. C., Kaufmann, P., & Cooper, P. E. (1996). Two genetically distinct populations of Fusarium oxysporum f. sp. lycopersici race 3 in the United States. Plant Disease, 80(12), 1336–1342.
Martens, M., Dawyndt, P., Coopman, R., Gillis, M., De Vos, P., & Willems, A. (2008). Advantages of multilocus sequence analysis for taxonomic studies: a case study using 10 housekeeping genes in the genus Ensifer (including former Sinorhizobium). International Journal of Systematic and Evolutionary Microbiology, 58, 200–214. https://doi.org/10.1099/ijs.0.65392-0
Meier-Kolthoff, J. P., & Göker, M. (2019). TYGS is an automated high-throughput platform for state-of-the-art genome-based taxonomy. Nature Communications, 10(1), 1–10. https://doi.org/10.1038/s41467-019-10210-3
Meier-Kolthoff, J. P., Auch, A. F., Klenk, H. P., & Göker, M. (2013). Genome sequence-based species delimitation with confidence intervals and improved distance functions. BMC Bioinformatics, 14(1), 1–14. https://doi.org/10.1186/1471-2105-14-60
Naureen, Z., Price, A. H., Hafeez, F. Y., & Roberts, M. R. (2009). Identification of rice blast disease-suppressing bacterial strains from the rhizosphere of rice grown in Pakistan. Crop Protection, 28(12), 1052–1060. https://doi.org/10.1016/j.cropro.2009.08.007
Panhwar, Q. A., Othman, R., Rahman, Z. A., Meon, S., & Ismail, M. R. (2012). Isolation and characterization of phosphate-solubilizing bacteria from aerobic rice. African Journal of Biotechnology, 11, 2711–2719.
Pérez-Miranda, S., Cabirol, N., George-Téllez, R., Zamudio-Rivera, L. S., & Fernández, F. J. (2007). O-CAS, a fast and universal method for siderophore detection. Journal of Microbiological Methods, 70(1), 127–131. https://doi.org/10.1016/j.mimet.2007.03.023
Richter, M., & Rosselló-Móra, R. (2009). Shifting the genomic gold standard for the prokaryotic species definition. Proceedings of the National Academy of Sciences, 106(45), 19126–19131. https://doi.org/10.1073/pnas.090641210
Richter, M., Rosselló-Móra, R., Oliver Glöckner, F., & Peplies, J. (2016). JSpeciesWS: A web server for prokaryotic species circumscription based on pairwise genome comparison. Bioinformatics, 32(6), 929–931. https://doi.org/10.1093/bioinformatics/btv681
Rong, X., & Huang, Y. (2010). Taxonomic evaluation of the Streptomyces griseus clade using multilocus sequence analysis and DNA-DNA hybridization, with proposal to combine 29 species and three subspecies as 11 genomic species. International Journal of Systematic and Evolutionary Microbiology, 60(Pt3), 696–703. https://doi.org/10.1099/ijs.0.012419-0. Epub 2009 Aug 5.
Saman, P., Kawicha, P., Sangdee, A., Wongpakdee, S., Rattanapolsan, L., Ponpang-Nga, P., Suwor, P., & Thanyasiriwat, T. (2022). Grafting Compatibility, Scion Growth, and Fusarium Wilt Disease Incidence of Intraspecific Grafted Tomato. Journal of Horticultural Research, 30(2), 95–104. https://doi.org/10.2478/johr-2022-0020
Sangdee, A., Kornphachara, S., & Srisawat, N. (2016). In vitro screening of antagonistic activity of soil Streptomyces against plant pathogenic fungi and assessment of its characters. Journal of Agricultural Technology, 12(1), 173–185.
Schwyn, B., & Neilands, J. B. (1987). Universal chemical assay for the detection and determination of siderophores. Analytical Biochemistry, 160(1), 47–56. https://doi.org/10.1016/0003-2697(87)90612-9
Sharma, P., & Thakur, D. (2020). Antimicrobial biosynthetic potential and diversity of culturable soil actinobacteria from forest ecosystems of Northeast India. Scientific Reports, 10(1), 1–18. https://doi.org/10.1038/s41598-020-60968-6
Shin, J. H., Fu, T., Park, K. H., & Kim, K. S. (2017). The effect of fungicides on mycelial growth and conidial germination of the ginseng root rot fungus. Cylindrocarpon Destructans. Mycobiology, 45(3), 220–225. https://doi.org/10.5941/MYCO.2017.45.3.220
Shrivastava, P., Kumar, R., & Yandigeri, M. S. (2017). In vitro biocontrol activity of halotolerant Streptomyces aureofaciens K20: A potent antagonist against Macrophomina phaseolina (Tassi) Goid. Saudi Journal of Biological Sciences, 24(1), 192–199. https://doi.org/10.1016/j.sjbs.2015.12.004
Siddiqui, Z. A. (2005). PGPR: prospective biocontrol agents of plant pathogens. In PGPR: biocontrol and biofertilization (pp. 111–142). Springer, Dordrecht. https://doi.org/10.1007/1-4020-4152-7_4
Sivalingam, P., Hong, K., Pote, J., & Prabakar, K. (2019). Extreme Environment Streptomyces: Potential Sources for New Antibacterial and Anticancer Drug Leads? International Journal of Microbiology, 2019:5283948. https://doi.org/10.1155/2019/5283948
Stackebrandt, E., & Ebers, J. (2006). Taxonomic parameters revisited: Tarnished gold standards. Microbiology Today, 33, 152–155.
Stulberg, E. R., Lozano, G. L., Morin, J. B., Park, H., Baraban, E. G., Mlot, C., Heffelfinger, C., Phillips, G. M., Rush, J. S., Phillips, A. J., Broderick, N. A., Thomas, M. G., Stabb, E. V., & Handelsman, J. (2016). Genomic and secondary metabolite analyses of Streptomyces sp. 2AW provide insight into the evolution of the cycloheximide pathway. Frontiers in microbiology, 7, 573. https://doi.org/10.3389/fmicb.2016.00573
Suárez-Moreno, Z. R., Vinchira-Villarraga, D. M., Vergara-Morales, D. I., Castellanos, L., Ramos, F. A., Guarnaccia, C., Degrassi, G., Venturi, V., & Moreno-Sarmiento, N. (2019). Plant-growth promotion and biocontrol properties of three Streptomyces spp. isolates to control bacterial rice pathogens. Frontiers in Microbiology, 10, 290.
Takamatsu, S., Lin, X., Nara, A., Komatsu, M., Cane, D. E., & Ikeda, H. (2011). Characterization of a silent sesquiterpenoid biosynthetic pathway in Streptomyces avermitilis controlling epi-isozizaene albaflavenone biosynthesis and isolation of a new oxidized epi-isozizaene metabolite. Microbial Biotechnology, 4(2), 184–191. https://doi.org/10.1111/j.1751-7915.2010.00209.x
Tamura, K., Stecher, G., & Kumar, S. (2021). MEGA11: Molecular evolutionary genetics analysis version 11. Molecular Biology and Evolution, 38(7), 3022–3027. https://doi.org/10.1093/molbev/msab120
Torguet, L., Zazurca, L., Martínez, G., Pons-Solé, G., Luque, J., & Miarnau, X. (2022). Evaluation of Fungicides and Application Strategies for the Management of the Red Leaf Blotch Disease of Almond. Horticulturae, 8(6), 501. https://doi.org/10.3390/horticulturae8060501
van Bergeijk, D. A., Terlouw, B. R., Medema, M. H., & van Wezel, G. P. (2020). Ecology and genomics of Actinobacteria: New concepts for natural product discovery. Nature Reviews Microbiology, 18(10), 546–558. https://doi.org/10.1038/s41579-020-0379-y
Vurukonda, S. S. K. P., Giovanardi, D., & Stefani, E. (2018). Plant growth promoting and biocontrol activity of Streptomyces spp. as endophytes. International journal of molecular sciences, 19(4), 952. https://doi.org/10.3390/ijms19040952
Weisburg, W. G., Barns, S. M., Pelletier, D. A., & Lane, D. J. (1991). 16S ribosomal DNA amplification for phylogenetic study. Journal of Bacteriology, 173(2), 697–703. https://doi.org/10.1128/jb.173.2.697-703.1991
Winding, A., Binnerup, S. J., & Pritchard, H. (2004). Non-target effects of bacterial biological control agents suppressing root pathogenic fungi. FEMS Microbiology Ecology, 47(2), 129–141. https://doi.org/10.1016/S0168-6496(03)00261-7
Acknowledgements
This research project was financially supported by Mahasarakham University. Rattana Pengproh graceful thanks the Ministry of Higher Education, Science, Research and Innovation. The authors also thank the Faculty of Natural Resources and Agro-Industry, Kasetsart University, Chalermphrakiat Sakon Nakhon Province Campus and the Department of Biology, Faculty of Science, Mahasarakham University for facilitating this project.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interest
The authors declare no conflict of interest.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Pengproh, R., Thanyasiriwat, T., Sangdee, K. et al. Antagonistic ability and genome mining of soil Streptomyces spp. against Fusarium oxysporum f. sp. lycopersici. Eur J Plant Pathol 167, 251–270 (2023). https://doi.org/10.1007/s10658-023-02698-9
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10658-023-02698-9