Abstract
Current evidence on COVID-19 prognostic models is inconsistent and clinical applicability remains controversial. We performed a systematic review to summarize and critically appraise the available studies that have developed, assessed and/or validated prognostic models of COVID-19 predicting health outcomes. We searched six bibliographic databases to identify published articles that investigated univariable and multivariable prognostic models predicting adverse outcomes in adult COVID-19 patients, including intensive care unit (ICU) admission, intubation, high-flow nasal therapy (HFNT), extracorporeal membrane oxygenation (ECMO) and mortality. We identified and assessed 314 eligible articles from more than 40 countries, with 152 of these studies presenting mortality, 66 progression to severe or critical illness, 35 mortality and ICU admission combined, 17 ICU admission only, while the remaining 44 studies reported prediction models for mechanical ventilation (MV) or a combination of multiple outcomes. The sample size of included studies varied from 11 to 7,704,171 participants, with a mean age ranging from 18 to 93 years. There were 353 prognostic models investigated, with area under the curve (AUC) ranging from 0.44 to 0.99. A great proportion of studies (61.5%, 193 out of 314) performed internal or external validation or replication. In 312 (99.4%) studies, prognostic models were reported to be at high risk of bias due to uncertainties and challenges surrounding methodological rigor, sampling, handling of missing data, failure to deal with overfitting and heterogeneous definitions of COVID-19 and severity outcomes. While several clinical prognostic models for COVID-19 have been described in the literature, they are limited in generalizability and/or applicability due to deficiencies in addressing fundamental statistical and methodological concerns. Future large, multi-centric and well-designed prognostic prospective studies are needed to clarify remaining uncertainties.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
As of December 2022, over 650 million cases of Corona Virus Disease 2019 (COVID-19), caused by the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), have been confirmed and over 6.6 million deaths globally were reported to the World Health Organization (WHO). To date, despite of vaccination efforts and other public health measures, viral transmission and therefore evolution, is a persisting challenge, with the numbers of confirmed cases still on the rise [1]. The clinical picture of COVID-19 infection is heterogeneous and ranges from asymptomatic or pre-asymptomatic phase, mild or moderate respiratory symptoms to severe viral pneumonia and acute respiratory distress syndrome, septic shock and/or multiple organ dysfunction requiring admission to intensive care unit (ICU), which might eventually lead to need for mechanical ventilation (MV), intubation, extracorporeal membrane oxygenation (ECMO) and death [2,3,4]. Early identification of COVID-19 patients at risk of critical illness is crucial for early identification of patients requiring urgent medical attention or who would benefit the most from treatment. In addition, early prediction of the disease course not only enables cost-effective allocation of health care resources, but potentially decreases fatality rates as well [5, 6]. The supply and demand of emergency department (ED) and ICU beds has created an imbalance, due to the large number of individuals affected by COVID-19, hence straining the available health care resources [7].
Demographics, comorbidities, physical examinations, laboratory parameters and imaging predictors have been tested in several studies and used to develop prognostic models for COVID-19. These models have been used to evaluate disease prognosis and to perform triage in case of scarce resources in some clinical settings [7]. Laboratory indicators including, but not limited to, lymphocyte and platelet count, creatinine, interleukin 6 (IL-6), procalcitonin (PCT), d-dimer, ferritin, lactate dehydrogenase (LDH), C-reactive protein (CRP), aspartate aminotransferase (AST), alanine aminotransferase (ALT), high-sensitivity troponin T (hs-TnT), albumin and creatine kinase (CK), have been identified as common predictors of poor outcomes in COVID-19 [8]. However, most of the reported models are at high risk of bias due to deficiencies in the methods used, definitions of COVID-19 (e.g., cases defined based on clinical features rather than on the result of laboratory diagnostic test for SARS-CoV-2) and the use of heterogeneous outcomes. Thus, there is an urgent need to comprehensively and critically assess the available literature and identify the best performing and methodologically rigorous prognostic models for COVID-19 progression.
With that in mind, we performed this systematic review to summarize and critically appraise the available studies that have developed, assessed and/or validated clinical prognostic models for COVID-19 to predict progression to severe or critical disease, ICU admission, need for MV, intubation, HFNT and mortality.
Methods
Data sources and search strategy
We conducted our systematic review following a recently published guide on performing systematic reviews and meta-analyses [9] and report based on the Preferred Reporting Items for Systematic Review and Meta-analyses (PRISMA) recommendations [10]. The protocol was registered with the international prospective register of systematic reviews (PROSPERO) with ID: CRD42021257478. We searched 6 electronic databases: Embase Ovid, Medline Ovid, Cochrane Central, Web of Science Core Collection, WHO COVID-19 Global literature on coronavirus disease and Google Scholar from inception to June 20th, 2022. To identify relevant records, we combined (a) COVID-19 related terms with (b) prognostic model-related terms, such as “risk prediction models”, “biomarkers”, and (c) outcome type: mortality, ICU admission, intubation, HFNT and ECMO. We performed our search with the assistance of an experienced medical librarian. We used EndNote to manage references. Details on the search strategy are provided in the Supplemental material: Appendix A.
Study selection
We used the following criteria in selecting studies for inclusion: (a) conducted in adult humans diagnosed with SARS-CoV-2 infection and/or COVID-19; (b) analyzed data from a prospective or retrospective cohort; (c) used a prognostic model to predict one or more adverse outcomes of COVID-19; (d) reported on the model’s predictive performance; and (e) published in a peer-reviewed journal. There was no restriction on publication year or language. We excluded case-reports, case–control and cross-sectional studies, dissertation abstracts, letters to the editor, conference proceedings, systematic reviews or meta-analyses, books, book chapters and animal studies. Two reviewers independently evaluated abstracts and full texts of the studies. Discrepancies between reviewers were resolved through a consensus or in consultation with a third independent reviewer.
Data extraction
Data from included studies were extracted independently in duplicate, based on various domains including the number of participants, predictors, outcomes, data analysis details and performance of the prediction model (including, but not limited to, AUC/c-index, sensitivity, specificity, positive and negative predictive values, likelihood ratio, accuracy, etc.). The extracted data were cross-checked by two reviewers and the complete data extraction form can be found in the Supplemental material, Tables 3 and 4.
Quality assessment
We used the PROBAST checklist to evaluate potential sources of bias and issues of individual studies that may affect the applicability of results in relation to the intended use of models. PROBAST includes 20 signaling questions across 4 domains (i.e., participants, predictors, outcome, and analysis). Two reviewers conducted this assessment independently. Any disagreements were handled by discussion and consensus. Following the PROBAST standards, if a prediction model evaluation was judged at low risk on all domains relating to bias and applicability, then the overall judgment was assessed as “low risk of bias (RoB)” or “low concern regarding applicability”. If an evaluation was judged as high RoB for at least 1 domain, it was considered as “high RoB” or “high concern regarding applicability”. If the prediction model evaluation was unclear in 1 or more domains and was rated as low in the remaining domains, it was judged as having “unclear RoB” or “unclear concern regarding applicability” [11]. Key information was organized by relevant domains in the Supplemental material, Table 5.
Data synthesis
For each study, we reported measures quantifying the performance of the prediction model and 95% confidence intervals (CI), when provided. Heterogeneity permitting, we sought to pool the results using either a fixed or random effects meta-analysis model to provide the pooled estimates and the corresponding 95% CIs.
Results
Literature search
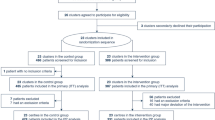
After excluding duplicates, 8,908 unique references were identified. Based on the initial screening of titles and abstracts, the full texts of 657 articles were retrieved and further evaluated. After full-text assessment, 343 studies were excluded due to inappropriate study design, no relevant outcome, unavailable full-text, pre-prints, irrelevance to research question, model performance not reported, or studies including human participants under 18 years. After examining full-texts, we identified 314 eligible articles, of which 152 (48.4%) presented mortality as the outcome [4, 5, 12,13,14,15,16,17,18,19,20,21,22,23,24,25,26,27,28,29,30,31,32,33,34,35,36,37,38,39,40,41,42,43,44,45,46,47,48,49,50,51,52,53,54,55,56,57,58,59,60,61,62,63,64,65,66,67,68,69,70,71,72,73,74,75,76,77,78,79,80,81,82,83,84,85,86,87,88,89,90,91,92,93,94,95,96,97,98,99,100,101,102,103,104,105,106,107,108,109,110,111,112,113,114,115,116,117,118,119,120,121,122,123,124,125,126,127,128,129,130,131,132,133,134,135,136,137,138,139,140,141,142,143,144,145,146,147,148,149,150,151,152,153,154,155,156,157,158,159,160], 66 (21.0%) concentrated on severity and/or critical illness [2, 161,162,163,164,165,166,167,168,169,170,171,172,173,174,175,176,177,178,179,180,181,182,183,184,185,186,187,188,189,190,191,192,193,194,195,196,197,198,199,200,201,202,203,204,205,206,207,208,209,210,211,212,213,214,215,216,217,218,219,220,221,222,223,224,225], 35 (11.1%) presented ICU admission and mortality combined [7, 226,227,228,229,230,231,232,233,234,235,236,237,238,239,240,241,242,243,244,245,246,247,248,249,250,251,252,253,254,255,256,257,258,259], 17 (5.4%) assessed ICU admission only [225, 260,261,262,263,264,265,266,267,268,269,270,271,272,273,274,275], 6 (1.9%) looked at mechanical ventilation only [276,277,278,279,280,281] and 38 (12.1%) assessed multiple combined outcomes [195, 282,283,284,285,286,287,288,289,290,291,292,293,294,295,296,297,298,299,300,301,302,303,304,305,306,307,308,309,310,311,312,313,314,315,316,317,318]. We present the results in the PRISMA flow chart Fig. 1. A meta-analysis was not feasible because of the large heterogeneity of study designs, measurement techniques, methods of analysis and handling of continuous variables, inclusion criteria, as well as reporting of different outcome measures.
Characteristics of the included studies
Of the 314 studies meeting eligibility criteria, 111 (35.4%) were conducted in Asia, 113 (36.0%) in Europe, 52 (16.6%) in North and Central America, 16 (5.1%) in South America, 7 (2.2%) in Africa, 1 (0.3%) in Australia and 14 (4.5%) were multi-national studies. The sample size ranged from 11 to 7,704,111 participants. Two hundred and seventy-five studies (87.6%) were based on retrospective study design, 36 (11.5%) on prospective study design and 3 (0.9%) on ambispective study design. The mean age ranged from 18 to 93 years with percentage of male participants ranging between 31.8–100%. Fifty-seven studies explored only model development, 26 performed only model validation, 167 developed and validated models internally and/or externally and 64 studies either assessed, analyzed, identified or compared prediction biomarkers.
The research population in the studies covered adult population (> 18 years old) presenting in the emergency room with clinical suspicion of COVID-19 or patients who were admitted to the hospital with laboratory confirmed SARS-CoV-2 infection by polymerase chain reaction (PCR) testing of nasopharyngeal samples. In 6 studies, the diagnosis was based on COVID-19 symptoms only. Of the 314 studies, 271 focused on hospitalized patients at baseline, 37 on hospitalized patients in the ICU, 5 focused on non-hospitalized patients and one study included both hospitalized patients in the ICU and non-hospitalized.
Out of 314 studies, 55 (17.5%) of the total studies adhered to transparent reporting of a multivariable prediction model for individual prognosis or diagnosis (TRIPOD) model evaluation guidelines. Across studies, multivariate logistic regression analysis was the most frequently used method of model development, followed by logistic regression, univariate logistic regression and machine learning analysis. We present the general characteristics of the eligible articles in the Supplementary material, Table 1.
Models predicting mortality
The most common outcome was mortality which was evaluated in 152 (48.4%) studies [4, 5, 12,13,14,15,16,17,18,19,20,21,22,23,24,25,26,27,28,29,30,31,32,33,34,35,36,37,38,39,40,41,42,43,44,45,46,47,48,49,50,51,52,53,54,55,56,57,58,59,60,61,62,63,64,65,66,67,68,69,70,71,72,73,74,75,76,77,78,79,80,81,82,83,84,85,86,87,88,89,90,91,92,93,94,95,96,97,98,99,100,101,102,103,104,105,106,107,108,109,110,111,112,113,114,115,116,117,118,119,120,121,122,123,124,125,126,127,128,129,130,131,132,133,134,135,136,137,138,139,140,141,142,143,144,145,146,147,148,149,150,151,152,153,154,155,156,157,158,159,160]. The population sample size ranged from 11 to 6,952,440 participants. The mean age ranged from 18 to 93 years, and the percentage of male participants from 31.8% to 100%. Among studies that specified and reported prediction time for mortality there was variation in reporting. In only 4 studies, prediction time was defined in a fixed time frame, i.e., within the time interval 8 to 30 days. The range of prediction time in the latter studies was between 24 h to 90 days. However, in some studies, prediction time was defined as time-to-event (i.e., death). The difference between fixed time frame and time-to-event outcomes, lies mainly in the fact that time-to-event reporting takes account of whether an event takes place and the time at which the event occurs, such that both the event and the timing of the event are important. In addition, it shows the differences duration of survival. Fixed timeframe reporting is commonly used when outcome measure data are collected for each participant and does not correspond to the overall duration of the study, but to a specified timeframe within which the outcome is most expected. By ‘prediction time’ we mean the timeframe which the model prespecified to assess its performance. Predictors encountered most frequently in the 152 studies that developed or validated mortality prediction models were increased age, sex, decreased oxygen saturation, elevated levels of CRP, blood urea nitrogen, body temperature, number of comorbidities, unconsciousness, white blood cells count, lymphocyte count, D-dimer level, platelets and pulse rate. Model performances assessed with AUC/ROC or c‐index were reported in 133 studies and ranged between 0.49 to 0.99. Additionally, sensitivity and specificity were reported in 73 studies ranging from 15.4 to 100% and 10.9 to 98.7%, respectively. Thirteen (8.6%) of the 152 included studies focused solely on model validation, whereas 83 (54.6%) combined development with internal and/or external validation. The best reported predictive performance belonged to a model developed in Boston, USA, and externally validated in Wuhan, China, with 375 participants. The development cohort, internal validation, and external validation’s respective AUC scores were 0.987, 0.999, and 0.992, respectively. The model was based on information acquired on admission, including age, lymphocyte count, d-dimer, CRP and creatinine (ALDCC) [123]. We present the detailed characteristics of the eligible articles based on mortality as outcome in the Supplementary material Table 2.
Models predicting severity or critical illness
Severity or critical illness in COVID-19 was reported in 66 (21.0%) [2, 161,162,163,164,165,166,167,168,169,170,171,172,173,174,175,176,177,178,179,180,181,182,183,184,185,186,187,188,189,190,191,192,193,194,195,196,197,198,199,200,201,202,203,204,205,206,207,208,209,210,211,212,213,214,215,216,217,218,219,220,221,222,223,224,225]. The definition of severity or critical outcome in COVID-19 varied across the studies. Some studies used the standard definition, which grades COVID-19 severity using the following criteria: (1) shortness of breath: respiratory rate > 30 breaths/min in the resting state; (2) pulse oxygen saturation < 93% or (3) arterial blood oxygen pressure (PaO2)/oxygen concentration (FiO2) < 300 mmHg. Criteria for critical patients includes: (1) respiratory failure requiring MV; (2) shock; (3) other multi-organ failure requiring ICU monitoring treatment. While other studies defined severity or critical illness as the occurrence of any of the following events: ICU admission, need for invasive mechanical ventilation (IMV) or death. We present the detailed definitions of COVID-19 severity or critical illness in the Supplemental material Table 6. Sample size ranged from 55 to 7,704,111 participants. The mean age ranged from 38.2 to 87.0 years with percentage male ranging from 41.4% to 77%. Prediction time defined as timeframe which the model pre-specified to assess its performance, i.e., from admission to the worsening severity or critical illness was different across studies, and the longest time of follow-up was 30 days. The most frequently encountered predictors were older age, sex, body temperature, number of comorbidities (cardiovascular disease (CVD), hypertension, diabetes), decreased oxygen saturation, elevated levels of CRP, blood urea nitrogen (BUN), body temperature, systolic blood pressure (SBP), neutrophil-to-lymphocyte ratio (NLR), white blood cells count (WBC), lymphocyte count and pulse rate. Model performances were assessed with AUC ranging from 0.57 to 0.99 in 60 studies, and sensitivity and specificity ranging from 7.1 to 100% and 19.5% − 100%, respectively. Thirty-nine (59.1%) of the 66 studies that were considered, combined development with internal and/or external validation, whereas 3 (4.5%) of the studies only focused on model validation. A combined machine learning model (Support Vector Machine (SVM), Gradient Boosted Decision Tree (GBDT), and Neural Network(NN)) based on procalcitonin, [T + B + NK cell] count, IL-6, CRP, IL-2-receptor, T-helper lymphocyte/T-suppressor lymphocyte as predictors of critical illness had the best reported predictive performance, with an AUC of 0.99. This model was developed in China with 450 participants (NN) [174]. We present detailed characteristics in the supplemental material Table 4.
Models predicting mortality and ICU admission
Thirty-five (11.1%) out of 314 studies reported mortality and ICU admission as outcome [7, 226,227,228,229,230,231,232,233,234,235,236,237,238,239,240,241,242,243,244,245,246,247,248,249,250,251,252,253,254,255,256,257,258,259]. Sample size in these studies ranged from 53 to 5,831 participants. The mean age differed from 43 to 82.2 years with percentage male ranging from 46.8% to 70.3%. Prediction time or the time from admission to ICU admission and death ranged from 24 h to 30 days. Common predictors were age, gender, pulse rate, albumin, WBC count, procalcitonin, LDH, CRP, ferritin, BUN, comorbidities and oxygen saturation among others. The AUC ranged from 0.63 to 0.98 in 32 studies, and sensitivity and specificity in 19 studies ranging from 10.5–98.7% from 41–100%, respectively. Eleven (31.4%) studies combined development with internal and/or external validation, whereas 4 (11.4%) of the studies only focused on model validation. Among these studies, the biomarker with the best predictive performance was CRP, showing an AUC of 0.975 [234]. We present detailed characteristics of the studies in the Supplemental material Table 4.
Models predicting ICU admission only
In total, there were only 17 (5.4%) studies which reported ICU admission as their outcome [225, 260,261,262,263,264,265,266,267,268,269,270,271,272,273,274,275]. Sample size in these studies varied from 67 to 4663 participants with a mean age ranging from 40 to 71.4 years and percentage male between 39.5% and 75.8%. Predictors reported were patients’ age, sex, presence of hypertension and diabetes, fever, short-ness of breath, serum glucose, AST, respiratory rate, NLR ratio, LDH, systolic blood pressure, CRP and fibrinogen. Model performances were assessed with AUC ranging from 0.44 to 0.97 in 15 studies, and sensitivity and specificity in 11 studies ranging from 30.2% to 92.4% and 45.5% to 99.7%, respectively.
Ten out of 17 studies (58.8%) provided combined development with internal and/or external validation, with the best reported predictive performance belonging to a model developed in Greece with 67 participants and an AUC of 0.97 based on patients’ gender, presence of hypertension and diabetes, fever, shortness of breath, serum glucose, AST, LDH, CRP and fibrinogen [261].We present detailed characteristics of the studies in the supplemental material Table 4.
Models predicting combined outcomes
Six (1.9%) of 314 studies reported models predicted MV only [195, 282,283,284,285,286,287,288,289,290,291,292,293,294,295,296,297,298,299,300,301,302,303,304,305,306,307,308,309,310,311,312,313,314,315,316,317,318], and 38 (12.1%) reported on a combination of outcomes (intubation, HFNT, ECMO, ICU admission and mortality) [195, 282,283,284,285,286,287,288,289,290,291,292,293,294,295,296,297,298,299,300,301,302,303,304,305,306,307,308,309,310,311,312,313,314,315,316,317,318]. Prediction time defined as timeframe which the model pre-specified to assess its performance, i.e., from admission to event outcomes, was different across studies, and the longest time of follow-up here was 30 days. AUC assessed in all 44 studies ranged from 0.53 to 0.94, and sensitivity and specificity ranging from 21.5% to 98.6% and 13.7% to 89.2%, respectively. A combination of model development with internal and/or external validation was provided by 24 (54.5%) out of 44 studies with an AUC of 0.94 for the two models with the best predictive performance [294, 316]. Additional information on prediction time, model performance and predictors are presented in Supplementary material Table 4.
Risk assessment
In the participants domain, 31/314 (9.9%) studies were rated as being at low RoB, 15/314 (4.8%) as unclear and 268/314 (85.4%) were at high RoB. High RoB in the participants domain mainly resulted from retrospective study design, in which data were generated from existing sources such as existing cohort studies or routine care registries.
In the predictors domain, 40/314 (13.1%) studies were rated as being at low RoB, 118/314 (37.6%) as unclear and 156/119 (49.7%) were at high RoB. The 40 studies with low RoB were rated as such given that the predictor assessment was conducted without prior knowledge of the outcome. However, most of the studies did not report information on predictor blinding, hence high or unclear RoB.
In the outcome domain, 53/314 (16.9%) studies were rated as being at low RoB, 157/314 (50.0%) as unclear and 104/314 (33.1%) as being at high RoB. High or unclear RoB was mainly due to the outcome being determined with prior knowledge of predictor information, leading to biased predictive performance. Another reason was lack of information on the time interval between predictor assessment and outcome determination in most of the studies.
In the analysis domain, 4/314 (1.3%) studies were rated as being at low RoB, 30/314 (9.6%) as unclear and 280/314 (89.2%%) as high RoB. Some of the reasons leading to studies being rated at high RoB were: (i) unreasonable number of participants with the outcome; (ii) inappropriate method of handling missing data; (iii) predictors selected based on univariable analysis prior to multivariable modelling and (iv) calibration and discrimination were not evaluated and no internal validation preformed.
Inappropriate method of handling missing data is related to the sampling, as a statistically valid analysis for a prognostic model which has appropriate mechanisms and assumptions for the missing data should be conducted to consider it at low risk of bias. The ‘appropriateness’ of handling missing values depends on the type of missing data, as well as the assumptions based on the reasons for the missing data. For instance, if the data that are missing are missing completely at random, the analysis is likely to be unbiased. Power may be lost in the design, but the estimated parameters are not biased by the absence of the data. However, if data are not missing at random, this might be problematic and there are specific methods that should be used to handle them. For instance, one way to obtain an unbiased estimate of the parameters in such a case is to model the missing data. The model may then be incorporated into a more complex one for estimating the missing values. Another way is to conduct a sensitivity analysis, or apply techniques like listwise or case deletion, pairwise deletion, mean substitution, regression imputation, last observation carried forward, maximum likelihood, expectation–maximization, multiple imputation, etc. Knight et al. (2022) [304] used multiple imputation to deal with missing data and examined heterogeneity in detail by NHS region, ethnicity and month of admission, and could serve as low RoB example.
Overall, the RoB assessment was rated to be at high or unclear RoB in 312 studies. This could be explained by shortcomings such as poor methodological quality, small sample size, poor handling of missing data, failure to deal with overfitting, definitions of COVID-19 based on clinical features rather than on the result of laboratory diagnostic test for SARSCoV-2 and its severity with studies using heterogeneous outcomes. Only two studies [82, 304], were considered to have a high level of methodological rigor with overall low RoB in all the domains. Knight et al., developed and validated the 4C Mortality score, which was rated as of good quality. This model includes 8 variables namely age, sex, number of comorbidities, respiratory rate, peripheral oxygen saturation, level of consciousness, urea level and CRP. It showed high discrimination for mortality with AUC of 0.77, (95% CI 0.76—0.77) in the validation cohort. The same 4C prognostic model was further validated in a large prospective cohort in UK and it showed AUCs of 0.78 (95% CI 0.77—0.78) and 0.76 (95% CI 0.75—0.77) for 4Cmortality and 4C Deterioration scores respectively.
According to the validation study, both the 4C Mortality and the 4C Deterioration scores can be used to stratify patients who have been admitted to the hospital with confirmed COVID-19. Both scores can also be used to make treatment decisions [82, 304].
Data availability
Only 14 (4.5%) of the 314 studies reviewed indicated the availability of data by either providing a link or where the data could be found. In a hundred and six (33.8%) studies, the authors stated that data are available upon request. The majority of the studies did not mention of the availability of the data.
Discussion
Fig. 2 To our knowledge, this is the most comprehensive systematic review of developed and/or validated clinical prognostic models for COVID-19. Our results indicate that only one predictive model was considered to be at low RoB, while the rest of the models were developed from studies suffering from many methodological issues including limitations in model development, presentation and being incompletely or inadequately reported. Thus, the polarized focus on limitations and methodological challenges that emerged from this report serves as a reminder that current prognostic models on COVID-19 severity, have limited applicability and require caution before implementation in routine care in the clinical setting. In addition, models were concentrated in specific regions, as they were developed and validated predominantly in China and Europe. Hence, there is a need to develop models tailored to other countries before generalization and application.
Contrary to the review done by Wynants et.al [319] which primarily focused on models for diagnosis and prognosis of COVID-19, our work exclusively focused on studies that either developed or validated models predicting COVID-19 severity and mortality. We included 314 studies that investigated either univariable or multivariable prognostic models that predict COVID-19 adverse outcomes, whereas Wynants et al. analyzed only multivariable related models and scoring systems. Thus, the studies included by Wynants et al. were limited in understanding single biomarkers that might have clinical utility in predicting COVID-19 severity. Additionally, in our research, all reviewed articles were peer-reviewed and published articles, omitting pre-prints for which there is also no guarantee that the information provided is supported by the data due to lack of formal peer review.
Eighty seven percent of the studies in this review are based on retrospective databases leading to lack of consistency in predictor and outcome measurement and deficiencies related to missing data. Of the included studies, only 36 studies were of prospective design. This indicates a high potential of model development and validation studies being appraised as being at high RoB, because the existing data sources, registries or existing cohorts were used. Data in the registries are usually collected for many purposes other than development of the prognostic models [320]. Thus, future studies using standardized and repeated measurements and prospective design can provide better answer to which extend prediction models can accurately predict COVID-19 severity.
In our review, only 55 (17.5%) studies adhered to the TRIPOD reporting guidelines. Similar to ours, previous reviews have shown that the quality of reporting in most of the articles describing model development or validation is relatively poor, and in the absence of detailed and transparent reporting of key study details, it is difficult for researchers to objectively judge the strengths and weaknesses of a prediction model study [321]. TRIPOD provides good reporting of the studies developing or validating prognostic models, thereby enabling the strength and weaknesses of a study to be revealed, hence facilitating its interpretation, and making it usable [11, 319].
Univariable and multivariable logistic regression analysis was the most used method of the prognostic model development in our review. Logistic regression is a widely used statistical method that allows for multivariable analysis and modelling of a binary dependent variable [322]. Other than logistic regression methods, machine learning methods such as decision tree (DT), gradient boosting decision trees (GB), support vector machine (SVM) and neural network (NN) were also applied to develop prediction models. Machine and artificial learning are becoming more common techniques due to increasing availability of large datasets. However, models developed based on machine learning and artificial intelligence must be carefully developed to reduce the risk of overfitting when data are sparse [11].
The overall sample size of the eligible studies in our review was ranging from 11 to 7,704,111 participants. However, by considering the commonly used rules of thumb to determine the sample size for prognostic models, it is evident that the sample sizes of some of the included studies in the review were relatively low. The number of EPV is the number of events divided by the number of predictor variables considered in developing the prediction model [323]. The rule of thumb recommends observation of at least 10 events per variable (EPV) and is based on estimates of stability of coefficient estimates for individual variables [324]. Future studies need to fulfil this criterion before seeking to validate these models externally and eventually predicting disease severity in a clinical scenario.
Our findings indicate that most studies do not provide information on availability of data, and among those studies providing information, only a minority provides information where the data can be found and accessed. Data availability and sharing is one of the cornerstones of quality science that allows reanalysis of data, reproducibility and merging of different dataset to address small sample size and other methodological issues. Thus, more work should be done in the future to address this issue, and facilitate access and exchange of data.
While we made a comprehensive search strategy, we do not exclude possibility of missing other relevant articles that reported regression results in abstract but the prognostic ability results only in full text. However, these studies very likely did not have as primary goal the generation or validation of a prognostic model, which was the primary interest of this review.
A future consideration for prognostic modelling studies should be the determination of the variant of the virus circulating, as certain variants, may be more severe than others, and may influence severity, adverse outcomes and death. For instance, in 2021, the Delta variant was identified and appeared to be more transmissible than the ancestral strain and also more severe. A study from the UK (n = 43,000 cases) showed that patients infected with Delta had twice the risk of hospitalization compared to those infected with Alpha, despite overall being younger [325].
Implications and recommendations
Future studies or researchers are recommended to use prospective longitudinal cohort design for prognostic model development and validation where the methods are prespecified and consistent. By adhering to consistent methods, participants data are systematically and validly recorded [11]. Another important aspect of prediction models is the internal and/or external validation process, which was done in 61.5% of studies included in this review. Future studies should therefore consider validating or comparing the existing models in different settings. To avoid models with biased selection of variables and inaccurate predictions, future studies are recommended to use large sample sizes or to report their justification for the choice of sample size [324]. Quality can be further improved by properly addressing the non-linear prognostic factors and missing values in studies. Authors are advised to follow the TRIPOD guidelines at the same time as they develop their predictive models in order to reduce their risk of bias [11, 321]. Considering that most included studies were found to be at high risk of bias, it is essential that future studies provide new and robust insight into the topic. Several prognostic models have been developed, but none is clearly superior nor accurately predicts deterioration or mortality to a great degree [326, 327]. The speed of symptom progression is not an accurate predictor of worse outcomes [328] and pre-intubation sequential organ assessment score has been shown to perform poorly as a predictor of death in patients with COVID-19 [329].
Additionally, researchers should consider well-designed prospective studies and biomarkers that could be easily assessed, readily available and are inexpensive to measure, to improve the clinical accessibility and applicability of the resulting models.
Finally, there is a need to build capacity and infrastructure to carry out research on prognostic tools that are potentially of benefit when applied in a clinical setting. Assessment of impact on decision-making should be critical when awarding grants for the development of predictive tools for quality improvement.
Example of good methods and reporting
Despite the general poor assessment on the quality of the articles, there were 2 articles that were appraised as low risk of bias, both by Knight et al. These articles used appropriate method and adhered to adequate reporting guidelines [82, 304].
The author included: (a) appropriate study design for prognostic models; (b) sufficient sample size to allow model development (event per variable (EPV) values of over 40); (c) reporting of the model method and coefficients of the final model. The 4C Mortality Score was found to have excellent discrimination and calibration in the validation cohort. It showed good applicability within the validation cohort and consistency across all performance measures. The score can accurately characterize the patients of high risk of death in hospital since it uses commonly available clinical observations, blood parameters and demographics at the time of hospital admission.
Conclusion
In this review, models predicting COVID-19 severity used comparable predictors, with different prediction performance. However, due to concerns in resolving statistical and methodological difficulties, the evidence is of relatively poor methodological rigor with limited generalizability and applicability. Future large, multi-center and well-designed prospective studies are needed for the development of predictive models for COVID-19 with clinical utility that can be applied to diverse populations. This requires a homogeneous definition of COVID-19 and outcomes and appropriate model selection methods to lead user-friendly models that can be externally validated.
Data availability
Data sharing is not applicable to this article as no datasets were generated or analyzed during the current study.
References
Tsang HF, et al. An update on COVID-19 pandemic: the epidemiology, pathogenesis, prevention and treatment strategies. Expert Rev Anti Infect Ther. 2021;19(7):877–88.
Bennouar S, et al. Development and validation of a laboratory risk score for the early prediction of COVID-19 severity and in-hospital mortality. Intensive Crit Care Nurs. 2021;64: 103012.
Eythorsson E, et al. Clinical spectrum of coronavirus disease 2019 in Iceland: population based cohort study. BMJ. 2020;371: m4529.
Aciksari G, et al. Evaluation of modified ATRIA risk score in predicting mortality in hospitalized patients with COVID-19. Am J Med Sci. 2021;362(6):553–61.
Weng Z, et al. ANDC: an early warning score to predict mortality risk for patients with coronavirus disease 2019. J Transl Med. 2020;18(1):328.
Kwok KO, et al. Epidemiology, clinical spectrum, viral kinetics and impact of COVID-19 in the Asia-Pacific region. Respirology. 2021;26(4):322–33.
Ak R, Kurt E, Bahadirli S. Comparison of 2 Risk prediction models specific for COVID-19: the Brescia-COVID respiratory severity scale versus the quick COVID-19 severity index. Disaster Med Public Health Prep. 2021;15(4):e46–50.
Malik P, et al. Biomarkers and outcomes of COVID-19 hospitalisations: systematic review and meta-analysis. BMJ Evid Based Med. 2021;26(3):107–8.
Muka T, et al. A 24-step guide on how to design, conduct, and successfully publish a systematic review and meta-analysis in medical research. Eur J Epidemiol. 2020;35(1):49–60.
Page MJ, et al. The PRISMA 2020 statement: an updated guideline for reporting systematic reviews. BMJ. 2021;372: n71.
Moons KGM, et al. PROBAST: a tool to assess risk of bias and applicability of prediction model studies: explanation and elaboration. Ann Intern Med. 2019;170(1):W1-w33.
Acar HC, et al. An easy-to-use nomogram for predicting in-hospital mortality risk in COVID-19: a retrospective cohort study in a university hospital. BMC Infect Dis. 2021;21(1):148.
Ahirwar AK, et al. The study of serum hsCRP, ferritin, IL-6 and plasma D-dimer in COVID-19: a retrospective study. Horm Mol Biol Clin Investig. 2022;43(3):337–44.
Al Abbasi B, et al. Cardiac troponin-I and COVID-19: a prognostic tool for in-hospital mortality. Cardiol Res. 2020;11(6):398–404.
Al Mutair A, et al. Clinical predictors of COVID-19 mortality among patients in intensive care units: a retrospective study. Int J Gen Med. 2021;14:3719–28.
Aletreby WT, et al. External validation of 4C ISARIC mortality score in critically ill COVID-19 patients from Saudi Arabia. Saudi J Med Med Sci. 2022;10(1):19–24.
Alfaro-Martinez JJ, et al. Generation and validation of in-hospital mortality prediction score in COVID-19 patients: Alba-score. Curr Med Res Opin. 2021;37(5):719–26.
Alhamar G, et al. Development of a clinical risk score to predict death in patients with COVID-19. Diabetes Metab Res Rev. 2022;38(5): e3526.
Alkaabi S, et al. A clinical risk score to predict in-hospital mortality in critically ill patients with COVID-19: a retrospective cohort study. BMJ Open. 2021;11(8): e048770.
Alkhasawneh RMR, et al. The predictive acccuracy for mortality of peripheral hematological markers ratios in mechanically ventilated COVID-19 patients. Arch Balkan Med Union. 2021;56(4):461–7.
Allahverdiyev S, et al. The neutrophil to lymphocyte ratio and in-hospital all-cause mortality in patients with COVID-19. Eur J Ther. 2020;26(3):251–6.
Altschul DJ, et al. A novel severity score to predict inpatient mortality in COVID-19 patients. Sci Rep. 2020;10(1):16726.
Andreano A, et al. Development of a multivariable model predicting mortality risk from comorbidities in an Italian cohort of 18,286 confirmed COVID-19 cases aged 40 years or older. Epidemiol Prev. 2021;45(1–2):100–9.
Araiza A, et al. The Ichikado CT score as a prognostic tool for coronavirus disease 2019 pneumonia: a retrospective cohort study. J Intensive Care. 2021;9(1):51.
Asaduzzaman MD, et al. Significance of hemogram-derived ratios for predicting in-hospital mortality in COVID-19: a multicenter study. Health Sci Rep. 2022;5(4): e663.
Asghar MS, et al. Poor prognostic biochemical markers predicting fatalities caused by COVID-19: a retrospective observational study from a developing country. Cureus. 2020;12(8): e9575.
Aygun H, Eraybar S. The role of emergency department triage early warning score (TREWS) and modified early warning score (MEWS) to predict in-hospital mortality in COVID-19 patients. Ir J Med Sci. 2022;191(3):997–1003.
Ayvat P, Kayhan Omeroglu S. Mortality estimation using APACHE and CT scores with stepwise linear regression method in COVID-19 intensive care unit: a retrospective study. Clin Imaging. 2022;88:4–8.
Bae J, et al. Predicting mechanical ventilation and mortality in COVID-19 using radiomics and deep learning on chest radiographs: a multi-institutional study. Diagnostics (Basel). 2021;11(10):1812.
Baikpour M, et al. Role of a chest X-ray severity score in a multivariable predictive model for mortality in patients with COVID-19: a single-center, retrospective study. J Clin Med. 2022;11(8):2157.
Banoei MM, et al. Machine-learning-based COVID-19 mortality prediction model and identification of patients at low and high risk of dying. Crit Care. 2021;25(1):328.
Bartoszko J, et al. Development of a repeated-measures predictive model and clinical risk score for mortality in ventilated COVID-19 patients. Can J Anaesth. 2022;69(3):343–52.
Beigmohammadi MT, et al. Mortality predictive value of APACHE II and SOFA scores in COVID-19 patients in the intensive care unit. Can Respir J. 2022;2022:5129314.
Bengelloun AK, et al. Usefulness of the CONUT index upon hospital admission as a potential prognostic indicator of COVID-19 health outcomes. Chin Med J (Engl). 2021;135(2):187–93.
Bertsimas D, et al. COVID-19 mortality risk assessment: an international multi-center study. PLoS ONE. 2020;15(12): e0243262.
Besutti G, et al. Imaging-based indices combining disease severity and time from disease onset to predict COVID-19 mortality: a cohort study. PLoS ONE. 2022;17(6): e0270111.
Besutti G, et al. The value of computed tomography in assessing the risk of death in COVID-19 patients presenting to the emergency room. Eur Radiol. 2021;31(12):9164–75.
Bezerra GF, et al. Urinary tubular biomarkers as predictors of death in critically ill patients with COVID-19. Biomark Med. 2022;16(9):681–92.
Bodolea C, et al. Nutritional risk assessment scores effectively predict mortality in critically Ill patients with severe COVID-19. Nutrients. 2022;14(10):2105.
Bradley J, et al. Pneumonia severity index and CURB-65 score are good predictors of mortality in hospitalized patients with SARS-CoV-2 community-acquired pneumonia. Chest. 2022;161(4):927–36.
Cai L, et al. Predictive nomogram for severe COVID-19 and identification of mortality-related immune features. J Allergy Clin Immunol Pract. 2021;9(1):177–84.
Cheng P, et al. Pneumonia scoring systems for severe COVID-19: which one is better. Virol J. 2021;18(1):33.
Chikhalkar B, et al. Assessment of national early warning score 2 as a tool to predict the outcome of COVID-19 patients on admission. Cureus. 2022;14(1): e21164.
Chou EH, et al. Development and validation of a prediction model for estimating one-month mortality of adult COVID-19 patients presenting at emergency department with suspected pneumonia: a multicenter analysis. Intern Emerg Med. 2022;17(3):805–14.
Churpek MM, et al. Machine learning prediction of death in critically Ill patients with coronavirus disease 2019. Crit Care Explor. 2021;3(8): e0515.
Cidade JP, et al. Predictive value of D-dimer in the clinical outcome of severe COVID19 patients: are we giving it too much credit? Clin Appl Thromb Hemost. 2022;28:10760296221079612.
Citu C, et al. Predictive value of blood coagulation parameters in poor outcomes in COVID-19 patients: a retrospective observational study in Romania. J Clin Med. 2022;11(10):2831.
Citu C, et al. Evaluation and comparison of the predictive value of 4C mortality score, NEWS, and CURB-65 in poor outcomes in COVID-19 patients: a retrospective study from a single center in Romania. Diagnostics (Basel). 2022;12(3):703.
Comoglu S, Kant A. Does the Charlson comorbidity index help predict the risk of death in COVID-19 patients? North Clin Istanb. 2022;9(2):117–21.
Cui X, et al. Establishment of prediction models for COVID-19 patients in different age groups based on random forest algorithm. QJM. 2022;114(11):795–801.
Ebell MH, et al. Development and validation of the COVID-NoLab and COVID-simplelab risk scores for prognosis in 6 US health systems. J Am Board Fam Med. 2021;34(Suppl):S127–35.
Ergenç H, et al. Procalcitonin/albumin ratio as a novel biomarker for predicting mortality in COVID-19. J Pioneering Med Sci. 11(1):3–7, 2022.
Falandry C, et al. Senior-COVID-rea cohort study: a geriatric prediction model of 30-day mortality in patients aged over 60 years in ICU for severe COVID-19. Aging Dis. 2022;13(2):614–23.
Fan X, et al. Scores based on neutrophil percentage and lactate dehydrogenase with or without oxygen saturation predict hospital mortality risk in severe COVID-19 patients. Virol J. 2021;18(1):67.
Fatemeh MT, et al. Identification of risk factors associated with mortality among patients with COVID-19 using random forest model: a historical cohort study. Acta Medica Iranica. 2021. https://doi.org/10.18502/acta.v59i8.7248.
Garrafa E, et al. Early prediction of in-hospital death of COVID-19 patients: a machine-learning model based on age, blood analyses, and chest x-ray score. Elife. 2021;10:e70640.
Golukhova EZ, et al. Transthoracic echocardiography-based prediction model of adverse event risk in patients with COVID-19. Pathophysiology. 2022;29(2):157–72.
Gordon AJ, et al. External validation of the 4C mortality score for hospitalised patients with COVID-19 in the RECOVER network. BMJ Open. 2022;12(4): e054700.
Gue YX, et al. Development of a novel risk score to predict mortality in patients admitted to hospital with COVID-19. Sci Rep. 2020;10(1):21379.
Gutierrez-Camacho JR, et al. Evaluation of the potential risk of mortality from SARS-CoV-2 infection in hospitalized patients according to the Charlson comorbidity index. Healthcare (Basel). 2022;10(2):362.
Haji Aghajani M, et al. Development of a scoring system for the prediction of in-hospital mortality among COVID-19 patients. Clin Epidemiol Glob Health. 2021;12: 100871.
Hajifathalian K, et al. Development and external validation of a prediction risk model for short-term mortality among hospitalized US COVID-19 patients: a proposal for the COVID-AID risk tool. PLoS One. 2020;15(9):e0239536.
Hassan S, et al. External validation of risk scores to predict in-hospital mortality in patients hospitalized due to coronavirus disease 2019. Eur J Intern Med. 2022;102:63–71.
He J, et al. Establishment of routine clinical indicators-based nomograms for predicting the mortality in patients With COVID-19. Front Med (Lausanne). 2021;8: 706380.
Heber S, et al. A model predicting mortality of hospitalized Covid-19 patients four days after admission: development, internal and temporal-external validation. Front Cell Infect Microbiol. 2021;11: 795026.
Hippisley-Cox J, et al. Risk prediction of covid-19 related death and hospital admission in adults after covid-19 vaccination: national prospective cohort study. BMJ. 2021;374: n2244.
Hohl CM, et al. The CCEDRRN COVID-19 mortality score to predict death among nonpalliative patients with COVID-19 presenting to emergency departments: a derivation and validation study. CMAJ Open. 2022;10(1):E90–9.
Hu C, et al. Early prediction of mortality risk among patients with severe COVID-19, using machine learning. Int J Epidemiol. 2021;49(6):1918–29.
Hu H, et al. Early prediction and identification for severe patients during the pandemic of COVID-19: a severe COVID-19 risk model constructed by multivariate logistic regression analysis. J Glob Health. 2020;10(2): 020510.
Hu H, Yao N, Qiu Y. Comparing rapid scoring systems in mortality prediction of critically Ill patients with novel coronavirus disease. Acad Emerg Med. 2020;27(6):461–8.
Huang CY, et al. The predictive and prognostic role of hematologic and biochemical parameters in the emergency department among coronavirus disease 2019 patients. Chin J Physiol. 2021;64(6):306–11.
Jain A, et al. Comparison of predictive ability of epidemiological factors, inflammatory biomarkers, and CT severity score for mortality in COVID-19. J Assoc Physicians India. 2021;69(8):11–2.
Jalalvand D, et al. Clinical, Laboratory, and Chest CT Scan Prognostic Factors for COVID-19 Mortality Cases. Trauma Monthly, 27:54-67. https://doi.org/10.30491/tm.2021.310669.1386.
Jamshidi E, et al. Symptom prediction and mortality risk calculation for COVID-19 using machine learning. Front Artif Intell. 2021;4: 673527.
Jiang M, et al. A biomarker-based age, biomarkers, clinical history, sex (ABCS)-mortality risk score for patients with coronavirus disease 2019. Ann Transl Med. 2021;9(3):230.
Kar S, et al. Multivariable mortality risk prediction using machine learning for COVID-19 patients at admission (AICOVID). Sci Rep. 2021;11(1):12801.
Khari S, et al. CURB-65, qSOFA, and SIRS criteria in predicting in-hospital mortality of critically Ill COVID-19 patients; a prognostic accuracy study. Arch Acad Emerg Med. 2022;10(1): e36.
Kibar Akilli I, et al. Comparison of pneumonia severity indices, qCSI, 4C-mortality score and qSOFA in predicting mortality in hospitalized patients with COVID-19 pneumonia. J Pers Med. 2022;12(5):801.
Kilercik M, et al. A new haematocytometric index: predicting severity and mortality risk value in COVID-19 patients. PLoS ONE. 2021;16(8): e0254073.
King JT Jr, et al. Development and validation of a 30-day mortality index based on pre-existing medical administrative data from 13,323 COVID-19 patients: the veterans health administration COVID-19 (VACO) Index. PLoS ONE. 2020;15(11): e0241825.
Klen R, et al. Development and evaluation of a machine learning-based in-hospital COVID-19 disease outcome predictor (CODOP): a multicontinental retrospective study. Elife. 2022. https://doi.org/10.7554/eLife.75985.
Knight SR, et al. Risk stratification of patients admitted to hospital with covid-19 using the ISARIC WHO clinical characterisation protocol: development and validation of the 4C mortality score. BMJ. 2020;370: m3339.
Kucuk B, et al. NUTRIC score is not superior to mNUTRIC score in prediction of mortality of COVID-19 patients. Int J Clin Pract. 2022;2022:1864776.
Laguna-Goya R, et al. IL-6-based mortality risk model for hospitalized patients with COVID-19. J Allergy Clin Immunol. 2020;146(4):799–807.
Laino ME, et al. An individualized algorithm to predict mortality in COVID-19 pneumonia: a machine learning based study. Arch Med Sci. 2022;18(3):587–95.
Lee DS, et al. Predictors of mortality among long-term care residents with SARS-CoV-2 infection. J Am Geriatr Soc. 2021;69(12):3377–88.
Leoni MLG, et al. Prediction of 28-day mortality in critically ill patients with COVID-19: development and internal validation of a clinical prediction model. PLoS ONE. 2021;16(7): e0254550.
Li G, et al. Uric acid as a prognostic factor and critical marker of COVID-19. Sci Rep. 2021;11(1):17791.
Li J, et al. Derivation and validation of a prognostic model for predicting in-hospital mortality in patients admitted with COVID-19 in Wuhan, China: the PLANS (platelet lymphocyte age neutrophil sex) model. BMC Infect Dis. 2020;20(1):959.
Li L, et al. Development and validation of a prognostic nomogram for predicting in-hospital mortality of COVID-19: a multicenter retrospective cohort study of 4086 cases in China. Aging (Albany NY). 2021;13(3):3176–89.
Li Y, et al. Development and validation of a two-step predictive risk stratification model for coronavirus disease 2019 in-hospital mortality: a multicenter retrospective cohort study. Front Med (Lausanne). 2022;9: 827261.
Liu H, et al. Development and validation of a risk score using complete blood count to predict in-hospital mortality in COVID-19 patients. Med (N Y). 2021;2(4):435-447e4.
Liu Q, et al. Laboratory findings and a combined multifactorial approach to predict death in critically ill patients with COVID-19: a retrospective study. Epidemiol Infect. 2020;148: e129.
Liu S, et al. Predictive performance of SOFA and qSOFA for in-hospital mortality in severe novel coronavirus disease. Am J Emerg Med. 2020;38(10):2074–80.
Lopez-Escobar A, et al. Risk score for predicting in-hospital mortality in COVID-19 (RIM Score). Diagnostics (Basel). 2021;11(4):596.
Lyons J, et al. Validating the QCOVID risk prediction algorithm for risk of mortality from COVID-19 in the adult population in Wales, UK. Int J Popul Data Sci. 2020;5(4):1697.
Ma X, et al. Characteristic of 523 COVID-19 in Henan province and a death prediction model. Front Public Health. 2020;8:475.
Ma X, et al. A nomogramic model based on clinical and laboratory parameters at admission for predicting the survival of COVID-19 patients. BMC Infect Dis. 2020;20(1):899.
Magro B, et al. Predicting in-hospital mortality from coronavirus disease 2019: a simple validated app for clinical use. PLoS ONE. 2021;16(1): e0245281.
Mahdavi M, et al. A machine learning based exploration of COVID-19 mortality risk. PLoS ONE. 2021;16(7): e0252384.
Marcolino MS, et al. ABC2-SPH risk score for in-hospital mortality in COVID-19 patients: development, external validation and comparison with other available scores. Int J Infect Dis. 2021;110:281–308.
Marincu I, et al. Predictive value of comorbid conditions for COVID-19 mortality. J Clin Med. 2021;10(12):2652.
Martin-Rodriguez F, et al. One-on-one comparison between qCSI and NEWS scores for mortality risk assessment in patients with COVID-19. Ann Med. 2022;54(1):646–54.
Morello F, et al. A 4C mortality score based dichotomic rule supports emergency department discharge of COVID-19 patients. Minerva Med. 2022;113(6):916–26.
Moulaei K, et al. Comparing machine learning algorithms for predicting COVID-19 mortality. BMC Med Inform Decis Mak. 2022;22(1):2.
Mousavi A, Rezaei S, Salamzadeh J, Mirzazadeh A, Peiravian F, Yousefi N. Value of laboratory tests in COVID-19 hospitalized patients for clinical decision-makers: a predictive model, using data mining approach. Iran Red Crescent Med J. 2021. https://doi.org/10.21203/rs.3.rs-56252/v1.
Munoz OM, et al. Validation of the PESI scale to predict in-hospital mortality in patients with pulmonary thromboembolism secondary to SARS CoV - 2 infection. Clin Appl Thromb Hemost. 2022;28:10760296221102940.
Murri R, et al. A machine-learning parsimonious multivariable predictive model of mortality risk in patients with Covid-19. Sci Rep. 2021;11(1):21136.
Najafi N, et al. The clinical course and prognostic factors of COVID-19 in patients with chronic kidney disease, a study in six centers. Iran J Kidney Dis. 2021;15(4):279–87.
Naser MN, et al. Risk factors, predictions, and progression of acute kidney injury in hospitalized COVID-19 patients: an observational retrospective cohort study. PLoS ONE. 2021;16(9): e0257253.
Nishikimi M, et al. Intubated COVID-19 predictive (ICOP) score for early mortality after intubation in patients with COVID-19. Sci Rep. 2021;11(1):21124.
Ocho K, et al. Clinical utility of 4C mortality scores among Japanese COVID-19 patients: a multicenter study. J Clin Med. 2022;11(3):821.
Ottenhoff MC, et al. Predicting mortality of individual patients with COVID-19: a multicentre Dutch cohort. BMJ Open. 2021;11(7): e047347.
Ozdemir IH, et al. Prognostic value of C-reactive protein/albumin ratio in hypertensive COVID-19 patients. Clin Exp Hypertens. 2021;43(7):683–9.
Ozdemir S, et al. Predictive ability of the MEWS, REMS, and RAPS in geriatric patients with SARS-CoV-2 infection in the emergency department. Disaster Med Public Health Prep. 2022. https://doi.org/10.1017/dmp.2022.107.
Ozger HS, et al. Serial measurement of cytokines strongly predict COVID-19 outcome. PLoS ONE. 2021;16(12): e0260623.
Pan D, et al. A predicting nomogram for mortality in patients with COVID-19. Front Public Health. 2020;8:461.
Pasculli P, et al. Chest computed tomography score, cycle threshold values and secondary infection in predicting COVID-19 mortality. New Microbiol. 2021;44(3):145–54.
Pigoga JL, Omer YO, Wallis LA. Derivation of a contextually-appropriate COVID-19 mortality scale for low-resource settings. Ann Glob Health. 2021;87(1):31.
Plecko D, et al. Rapid evaluation of coronavirus illness severity (RECOILS) in intensive care: development and validation of a prognostic tool for in-hospital mortality. Acta Anaesthesiol Scand. 2022;66(1):65–75.
Ponce D, et al. Development of a prediction score for in-hospital mortality in COVID-19 patients with acute kidney injury: a machine learning approach. Sci Rep. 2021;11(1):24439.
Quanjel MJR, et al. Replication of a mortality prediction model in Dutch patients with COVID-19. Nat Mach Intell. 2020;3(1):23–4.
Rahman T, et al. Mortality prediction utilizing blood biomarkers to predict the severity of COVID-19 using machine learning technique. Diagnostics (Basel). 2021;11(9):1592.
Rahman T, et al. Development and validation of an early scoring system for prediction of disease severity in COVID-19 using complete blood count parameters. IEEE Access. 2021;9:120422–41.
Raschke RA, et al. COVID-19 time of intubation mortality evaluation (C-TIME): a system for predicting mortality of patients with COVID-19 pneumonia at the time they require mechanical ventilation. PLoS ONE. 2022;17(7): e0270193.
Reina Reina A, et al. Machine learning model from a Spanish cohort for prediction of SARS-COV-2 mortality risk and critical patients. Sci Rep. 2022;12(1):5723.
Riley JM, et al. External validation of the COVID-19 4C mortality score in an urban United States cohort. Am J Med Sci. 2022;364(4):409–13.
Riva G, et al. Monocyte distribution Width (MDW) as novel inflammatory marker with prognostic significance in COVID-19 patients. Sci Rep. 2021;11(1):12716.
Rozenbaum D, et al. Personalized prediction of hospital mortality in COVID-19-positive patients. Mayo Clin Proc Innov Qual Outcomes. 2021;5(4):795–801.
Ruscica M, et al. Prognostic parameters of in-hospital mortality in COVID-19 patients-An Italian experience. Eur J Clin Invest. 2021;51(9): e13629.
Satici C, et al. Performance of pneumonia severity index and CURB-65 in predicting 30-day mortality in patients with COVID-19. Int J Infect Dis. 2020;98:84–9.
Selcuk M, et al. Comparison of D-dimer level measured on the third day of hospitalization with admission D-dimer level in predicting in-hospital mortality in COVID-19 patients. Medeni Med J. 2021;36(1):1–6.
Shanbehzadeh M, Nopour R, Kazemi-Arpanahi H. Design of an artificial neural network to predict mortality among COVID-19 patients. Inform Med Unlocked. 2022;31: 100983.
Shang Y, et al. Scoring systems for predicting mortality for severe patients with COVID-19. EClinicalMedicine. 2020;24: 100426.
Singh S, Singh K. Blood urea nitrogen/albumin ratio and mortality risk in patients with COVID-19. Indian J Crit Care Med. 2022;26(5):626–31.
Sosa FA, et al. Lung ultrasound as a predictor of mortality of patients with COVID-19. J Bras Pneumol. 2021;47(4): e20210092.
Soto-Mota A, et al. The low-harm score for predicting mortality in patients diagnosed with COVID-19: a multicentric validation study. J Am Coll Emerg Physicians Open. 2020;1(6):1436–43.
Stachel A, et al. Development and validation of a machine learning model to predict mortality risk in patients with COVID-19. BMJ Health Care Inform. 2021;28(1):e100235.
Surme S, et al. Novel biomarker-based score (SAD-60) for predicting mortality in patients with COVID-19 pneumonia: a multicenter retrospective cohort of 1013 patients. Biomark Med. 2022;16(8):577–88.
Tanboga IH, et al. Development and validation of clinical prediction model to estimate the probability of death in hospitalized patients with COVID-19: insights from a nationwide database. J Med Virol. 2021;93(5):3015–22.
Tezza F, et al. Predicting in-hospital mortality of patients with COVID-19 using machine learning techniques. J Pers Med. 2021;11(5):343.
Timpau AS, et al. Mortality predictors in severe COVID-19 patients from an East European tertiary center: a never-ending challenge for a no happy ending pandemic. J Clin Med. 2021;11(1):58.
Valente Silva B, et al. Prognostic accuracy of the modified CHA2DS2-VASc score in COVID-19 patients admitted to the emergency department due to clinical worsening. Acta Med Port. 2022;35(6):433–42.
van de Leur RR, et al. Electrocardiogram-based mortality prediction in patients with COVID-19 using machine learning. Neth Heart J. 2022;30(6):312–8.
Vicka V, et al. Comparison of mortality risk evaluation tools efficacy in critically ill COVID-19 patients. BMC Infect Dis. 2021;21(1):1173.
Vieira MLC, et al. A risk score for predicting death in COVID-19 in-hospital infection: a Brazilian single-center study. J Clin Ultrasound. 2022;50(5):604–10.
Wang L, et al. The utility of MEWS for predicting the mortality in the elderly adults with COVID-19: a retrospective cohort study with comparison to other predictive clinical scores. PeerJ. 2020;8: e10018.
Wang X, et al. Ratios of neutrophil-to-lymphocyte and platelet-to-lymphocyte predict all-cause mortality in inpatients with coronavirus disease 2019 (COVID-19): a retrospective cohort study in a single medical centre. Epidemiol Infect. 2020;148: e211.
Webb BJ, et al. Simple scoring tool to estimate risk of hospitalization and mortality in ambulatory and emergency department patients with COVID-19. PLoS ONE. 2022;17(3): e0261508.
Wirth A, et al. External validation of the modified 4C deterioration model and 4C mortality score for COVID-19 patients in a Swiss Tertiary Hospital. Diagn (Basel). 2022;12(5):1129.
Wongvibulsin S, et al. Development of severe COVID-19 adaptive risk predictor (SCARP), a calculator to predict severe disease or death in hospitalized patients With COVID-19. Ann Intern Med. 2021;174(6):777–85.
Yang Q, et al. Clinical characteristics and a decision tree model to predict death outcome in severe COVID-19 patients. BMC Infect Dis. 2021;21(1):783.
Yang Y, et al. Nomogram for prediction of fatal outcome in patients with severe COVID-19: a multicenter study. Mil Med Res. 2021;8(1):21.
Yilmaz E, et al.Which is better in predicting mortality in patients hospitalized for COVID-19: CURB-65 score versus physicians' gestalt. Southern Clinics of Istanbul Eurasia. 2021;32(3).
Yu JS, et al. Myoglobin offers higher accuracy than other cardiac-specific biomarkers for the prognosis of COVID-19. Front Cardiovasc Med. 2021;8: 686328.
Yuan Y, et al. Development and validation of a prognostic risk score system for COVID-19 Inpatients: a multi-center retrospective study in China. Eng (Beijing). 2022;8:116–21.
Zayed NE, Bessar MA, Lutfy S. CO-RADS versus CT-SS scores in predicting severe COVID-19 patients: retrospective comparative study. Egypt J Bronchol. 2021;15(1):1–10.
Zeng Z, et al. Development and validation of a simple-to-use nomogram to predict the deterioration and survival of patients with COVID-19. BMC Infect Dis. 2021;21(1):356.
Zhang S, et al. Development and validation of a risk factor-based system to predict short-term survival in adult hospitalized patients with COVID-19: a multicenter, retrospective, cohort study. Crit Care. 2020;24(1):438.
Zou X, et al. Acute physiology and chronic health evaluation II score as a predictor of hospital mortality in patients of coronavirus disease 2019. Crit Care Med. 2020;48(8):e657–65.
Ageno W, et al. Clinical risk scores for the early prediction of severe outcomes in patients hospitalized for COVID-19. Intern Emerg Med. 2021;16(4):989–96.
An H, et al. Inflammation/coagulopathy/immunology responsive index predicts poor COVID-19 prognosis. Front Cell Infect Microbiol. 2022;12: 807332.
Assal HH, et al. Predictors of severity and mortality in COVID-19 patients. Egypt J Bronchol. 2022;16(1):1–9.
Bello-Chavolla OY, et al. Validation and repurposing of the MSL-COVID-19 score for prediction of severe COVID-19 using simple clinical predictors in a triage setting: the nutri-CoV score. PLoS ONE. 2020;15(12): e0244051.
Bennett TD, et al. Clinical characterization and prediction of clinical severity of SARS-CoV-2 infection among US adults using data from the US national COVID cohort collaborative. JAMA Netw Open. 2021;4(7): e2116901.
Bennouar S, et al. Usefulness of biological markers in the early prediction of corona virus disease-2019 severity. Scand J Clin Lab Invest. 2020;80(8):611–8.
Boero E, et al. The COVID-19 worsening score (COWS)-a predictive bedside tool for critical illness. Echocardiography. 2021;38(2):207–16.
Chang Y, et al. Severe versus common COVID-19: an early warning nomogram model. Aging (Albany NY). 2022;14(2):544–56.
Chen W, et al. Development and validation of a clinical prediction model to estimate the risk of critical patients with COVID-19. J Med Virol. 2022;94(3):1104–14.
Chen Y, et al. CANPT score: a tool to predict severe COVID-19 on admission. Front Med (Lausanne). 2021;8: 608107.
De Socio GV, et al. National early warning score 2 (NEWS2) better predicts critical coronavirus disease 2019 (COVID-19) illness than COVID-GRAM, a multi-centre study. Infection. 2021;49(5):1033–8.
Erturk Sengel B, et al. Application of CALL score for prediction of progression risk in patients with COVID-19 at university hospital in Turkey. Int J Clin Pract. 2021;75(10): e14642.
Fernandes FT, et al. A multipurpose machine learning approach to predict COVID-19 negative prognosis in Sao Paulo, Brazil. Sci Rep. 2021;11(1):3343.
Gao Y, et al. Development and validation of an online model to predict critical COVID-19 with immune-inflammatory parameters. J Intensive Care. 2021;9(1):19.
Gómez LC, et al. Predictive model of severity in SARS CoV-2 patients at hospital admission using blood-related parameters. Ejifcc. 2021;32(2):255–64.
Gurusamy DS, et al. Role of neutrophil lymphocyte ratio as a prognostic biomarker in COVID-19 infection in indian patients: a single centre observational study from South India. J Clin Diagn Res. 2021;15(6):32–6.
Haimovich AD, et al. Development and validation of the quick COVID-19 severity index: a prognostic tool for early clinical decompensation. Ann Emerg Med. 2020;76(4):442–53.
Han Y, et al. Artificial intelligence computed tomography helps evaluate the severity of COVID-19 patients: a retrospective study. World J Emerg Med. 2022;13(2):91–7.
Huang J, et al. Indicators and prediction models for the severity of Covid-19. Int J Clin Pract. 2021;75(10): e14571.
Jiang J, et al. Development and validation of a predictive nomogram with age and laboratory findings for severe COVID-19 in Hunan Province. China Ther Clin Risk Manag. 2022;18:579–91.
Lee JY, et al. A risk scoring system to predict progression to severe pneumonia in patients with Covid-19. Sci Rep. 2022;12(1):5390.
Leyderman IN, et al. Prognostic value of serum albumin and urea nitrogen excretion in COVID-19 ICU patients: a single-center, prospective, cohort study. Ann Crit Care. 2021;3:61–8.
Li S, et al. Predictive value of chest CT scoring in COVID-19 patients in Wuhan, China: a retrospective cohort study. Respir Med. 2021;176: 106271.
Li XL, et al. Development and validation of a nomogram for predicting the disease progression of nonsevere coronavirus disease 2019. J Transl Int Med. 2021;9(2):131–42.
Liang W, et al. Development and validation of a clinical risk score to predict the occurrence of critical illness in hospitalized patients with COVID-19. JAMA Intern Med. 2020;180(8):1081–9.
Liu J, et al. Development and validation of a prediction model for early identification of critically ill elderly COVID-19 patients. Aging (Albany NY). 2020;12(19):18822–32.
Liu J, et al. The clinical course and prognostic factors of severe COVID-19 in Wuhan, China: a retrospective case-control study. Medicine (Baltimore). 2021;100(8): e23996.
Liu L, et al. Early prediction model for progression and prognosis of severe patients with coronavirus disease 2019. Medicine (Baltimore). 2021;100(8): e24901.
Liu Q, et al. Machine learning models for predicting critical illness risk in hospitalized patients with COVID-19 pneumonia. J Thorac Dis. 2021;13(2):1215–29.
Ma K, et al. Development and validation of a new prognostic scoring system for COVID-19. Jpn J Infect Dis. 2021;74(4):359–66.
Marcos M, et al. Development of a severity of disease score and classification model by machine learning for hospitalized COVID-19 patients. PLoS ONE. 2021;16(4): e0240200.
Monterde D, et al. Performance of three measures of comorbidity in predicting critical COVID-19: a retrospective analysis of 4607 hospitalized patients. Risk Manag Healthc Policy. 2021;14:4729–37.
Muto Y, et al. Predictive model for the development of critical coronavirus disease 2019 and its risk factors among patients in Japan. Respir Investig. 2021;59(6):804–9.
Myrstad M, et al. National early warning score 2 (NEWS2) on admission predicts severe disease and in-hospital mortality from Covid-19 - a prospective cohort study. Scand J Trauma Resusc Emerg Med. 2020;28(1):66.
Nádasdi Á, et al. Decreased circulating dipeptidyl peptidase-4 enzyme activity is prognostic for severe outcomes in COVID-19 inpatients. Biomark Med. 2022;16(5):317–30.
Nuevo-Ortega P, et al. Prognosis of COVID-19 pneumonia can be early predicted combining age-adjusted Charlson comorbidity index, CRB score and baseline oxygen saturation. Sci Rep. 2022;12(1):2367.
Patel M, et al. Utility of the ROX index in predicting intubation for patients with COVID-19-related hypoxemic respiratory failure receiving high-flow nasal therapy: retrospective cohort study. JMIRx Med. 2021;2(3): e29062.
Peng H, et al. Incubation period, clinical and lung CT features for early prediction of COVID-19 deterioration: development and internal verification of a risk model. BMC Pulm Med. 2022;22(1):188.
Prower E, et al. The ROX index has greater predictive validity than NEWS2 for deterioration in Covid-19. EClinicalMedicine. 2021;35: 100828.
Purkayastha S, et al. Machine learning-based prediction of COVID-19 severity and progression to critical illness using CT imaging and clinical data. Korean J Radiol. 2021;22(7):1213–24.
Rinderknecht MD, Klopfenstein Y. Predicting critical state after COVID-19 diagnosis: model development using a large US electronic health record dataset. NPJ Digit Med. 2021;4(1):113.
Schalekamp S, et al. Model-based prediction of critical illness in hospitalized patients with COVID-19. Radiology. 2021;298(1):E46–54.
Schoning V, et al. Development and validation of a prognostic COVID-19 severity assessment (COSA) score and machine learning models for patient triage at a tertiary hospital. J Transl Med. 2021;19(1):56.
Shalmon T, et al. Predefined and data driven CT densitometric features predict critical illness and hospital length of stay in COVID-19 patients. Sci Rep. 2022;12(1):8143.
Shankar V, et al. Development and validation of prognostic scoring system for COVID-19 severity in South India. Ir J Med Sci. 2022;191:2823–31.
Shi H, et al. CT-based radiomic nomogram for predicting the severity of patients with COVID-19. Eur J Med Res. 2022;27(1):13.
Shi Y, et al. Validation of pneumonia prognostic scores in a statewide cohort of hospitalised patients with COVID-19. Int J Clin Pract. 2021;75(3): e13926.
Su Y, et al. Prognostic accuracy of early warning scores for clinical deterioration in patients with COVID-19. Front Med (Lausanne). 2020;7: 624255.
Tang F, et al. A nomogram prediction of outcome in patients with COVID-19 based on individual characteristics incorporating immune response-related indicators. J Med Virol. 2022;94(1):131–40.
Tu C, et al. Establishment of a clinical nomogram model to predict the progression of COVID-19 to severe disease. Ther Clin Risk Manag. 2021;17:553–61.
Ucan ES, et al. Pneumonia severity indices predict prognosis in coronavirus disease-2019. Respir Med Res. 2021;79: 100826.
Vela E, et al. Development and validation of a population-based risk stratification model for severe COVID-19 in the general population. Sci Rep. 2022;12(1):3277.
Wong KC, et al. Uncovering clinical risk factors and predicting severe COVID-19 cases using UK biobank data: machine learning approach. JMIR Public Health Surveill. 2021;7(9): e29544.
Woo SH, et al. Development and validation of a web-based severe COVID-19 risk prediction model. Am J Med Sci. 2021;362(4):355–62.
Wu G, et al. Development of a clinical decision support system for severity risk prediction and triage of COVID-19 patients at hospital admission: an international multicentre study. Eur Respir J. 2020;56(2):2001104.
Xiao LS, et al. Development and validation of a deep learning-based model using computed tomography imaging for predicting disease severity of coronavirus disease 2019. Front Bioeng Biotechnol. 2020;8:898.
Xiong Y, et al. Comparing different machine learning techniques for predicting COVID-19 severity. Infect Dis Poverty. 2022;11(1):19.
Xu F, et al. Prediction of disease progression of COVID-19 based upon machine learning. Int J Gen Med. 2021;14:1589–98.
Xu J, et al. A predictive score for progression of COVID-19 in hospitalized persons: a cohort study. NPJ Prim Care Respir Med. 2021;31(1):33.
Yao Z, et al. Construction and validation of a machine learning-based nomogram: a tool to predict the risk of getting severe coronavirus disease 2019 (COVID-19). Immun Inflamm Dis. 2021;9(2):595–607.
Yu Y, et al. Novel biomarkers for the prediction of COVID-19 progression a retrospective, multi-center cohort study. Virulence. 2020;11(1):1569–81.
Zhang B, et al. Clinical utility of a nomogram for predicting 30-days poor outcome in hospitalized patients with COVID-19: multicenter external validation and decision curve analysis. Front Med (Lausanne). 2020;7: 590460.
Zhang H, et al. A nomogram predicting the severity of COVID-19 based on initial clinical and radiologic characteristics. Future Virol. 2022;17(4):221–9.
Zhao Q, et al. Early predictors of severe COVID-19 among hospitalized patients. J Clin Lab Anal. 2022;36(2): e24177.
Zhou Y, et al. Development and validation a nomogram for predicting the risk of severe COVID-19: a multi-center study in Sichuan, China. PLoS ONE. 2020;15(5): e0233328.
Adderley NJ, et al. Development and external validation of prognostic models for COVID-19 to support risk stratification in secondary care. BMJ Open. 2022;12(1): e049506.
Aguadero V, et al. Prognostic value of serum lactate dehydrogenase in hospitalized patients with Covid-19. Revista Romana de Medicina de Laborator. 2021;29(2):131–41.
Ahmed A, et al. Utility of severity assessment tools in COVID-19 pneumonia: a multicentre observational study. Clin Med (Lond). 2022;22(1):63–70.
Ak R, Doganay F. Comparison of four different threshold values of shock index in predicting mortality of COVID-19 patients. Disaster Med Public Health Prep. 2021. https://doi.org/10.1017/dmp.2021.374.
Arnold DT, et al. Predicting outcomes of COVID-19 from admission biomarkers: a prospective UK cohort study. Emerg Med J. 2021;38(7):543–8.
Asmarawati TP, et al. Predictive value of sequential organ failure assessment, quick sequential organ failure assessment, acute physiology and chronic health evaluation II, and new early warning signs scores estimate mortality of COVID-19 patients requiring intensive care unit. Indian J Crit Care Med. 2022;26(4):464–71.
Aznar-Gimeno R, et al. A clinical decision web to predict ICU admission or death for patients hospitalised with COVID-19 using machine learning algorithms. Int J Environ Res Public Health. 2021;18(16):8677.
Brook R, et al. Risk factors and early prediction of clinical deterioration and mortality in adult COVID-19 inpatients: an Australian tertiary hospital experience. Intern Med J. 2022;52(4):550–8.
Ceci FM, et al. Early routine biomarkers of SARS-CoV-2 morbidity and mortality: outcomes from an emergency section. Diagnostics (Basel). 2022;12(1):176.
Covino M, et al. Predicting intensive care unit admission and death for COVID-19 patients in the emergency department using early warning scores. Resuscitation. 2020;156:84–91.
Ganesan R, et al. Mortality prediction of COVID-19 patients at intensive care unit admission. Cureus. 2021;13(11): e19690.
Garcia Clemente MM, et al. Assessment of risk scores in Covid-19. Int J Clin Pract. 2021;75(12): e13705.
Hormanstorfer M, et al. Prognostic value of static and dynamic biomarkers in COVID-19 patients: a prospective cohort study. Rev Esp Quimioter. 2021;34(4):308–14.
Jibril H, et al. Predicting development of critical illness in patients with covid-19 presenting to a tertiary care hospital in a developing country. J Ayub Med Coll Abbottabad. 2022;34(2):256–62.
Kurt E, Bahadirli S. The usefulness of shock index and modified shock index in predicting the outcome of COVID-19 patients. Disaster Med Public Health Prep. 2022;16(4):1558–63.
Lazar Neto F, et al. Community-acquired pneumonia severity assessment tools in patients hospitalized with COVID-19: a validation and clinical applicability study. Clin Microbiol Infect. 2021;27(7):1037e1–8.
Levine DM, et al. Derivation of a Clinical risk score to predict 14-day occurrence of hypoxia, ICU admission, and death among patients with coronavirus disease 2019. J Gen Intern Med. 2021;36(3):730–7.
Li X, et al. Deep learning prediction of likelihood of ICU admission and mortality in COVID-19 patients using clinical variables. PeerJ. 2020;8: e10337.
Munera N, et al. A novel model to predict severe COVID-19 and mortality using an artificial intelligence algorithm to interpret chest radiographs and clinical variables. ERJ Open Res. 2022. https://doi.org/10.1183/23120541.00010-2022.
Pokeerbux MR, et al. National early warning score to predict intensive care unit transfer and mortality in COVID-19 in a French cohort. Int J Clin Pract. 2021;75(6): e14121.
Ponsford MJ, et al. Examining the utility of extended laboratory panel testing in the emergency department for risk stratification of patients with COVID-19: a single-centre retrospective service evaluation. J Clin Pathol. 2022;75(4):255–62.
Prasetya IB, et al. Prognostic value of inflammatory markers in patients with COVID-19 in Indonesia. Clin Epidemiol Glob Health. 2021;11: 100803.
Rasyid H, et al. Impact of age to ferritin and neutrophil-lymphocyte ratio as biomarkers for intensive care requirement and mortality risk in COVID-19 patients in Makassar, Indonesia. Physiol Rep. 2021;9(10): e14876.
Regolo M, et al. Neutrophil-to-lymphocyte ratio (NLR) Is a promising predictor of mortality and admission to intensive care unit of COVID-19 patients. J Clin Med. 2022;11(8):2235.
Rizzi M, et al. Prognostic markers in hospitalized COVID-19 patients: the role of IP-10 and C-reactive protein. Dis Markers. 2022;2022:3528312.
Rodriguez-Nava G, et al. Performance of the quick COVID-19 severity index and the Brescia-COVID respiratory severity scale in hospitalized patients with COVID-19 in a community hospital setting. Int J Infect Dis. 2021;102:571–6.
Shi S, et al. Prediction of adverse clinical outcomes in patients with coronavirus disease 2019. J Clin Lab Anal. 2021;35(1): e23598.
Subudhi S, et al. Comparing machine learning algorithms for predicting ICU admission and mortality in COVID-19. NPJ Digit Med. 2021;4(1):87.
Usul E. The role of the quick sequential organ failure assessment Score (qSOFA) in the Pre-hospitalization prediction of Covid19 prognosis. Erciyes Med J. 2021;43(5):494–9.
van Dam PM, et al. Validating the RISE UP score for predicting prognosis in patients with COVID-19 in the emergency department: a retrospective study. BMJ Open. 2021;11(2): e045141.
van Klaveren D, et al. COVID outcome prediction in the emergency department (COPE): using retrospective Dutch hospital data to develop simple and valid models for predicting mortality and need for intensive care unit admission in patients who present at the emergency department with suspected COVID-19. BMJ Open. 2021;11(9): e051468.
Wilfong EM, et al. Severity of illness scores at presentation predict ICU admission and mortality in COVID-19. J Emerg Crit Care Med. 2021. https://doi.org/10.21037/jeccm-20-92.
Zahedin K, et al. Neutrophil-to-lymphocyte ratio cut-off point for COVID-19 mortality: a retrospective study. Acta Medica Iranica. 2022;60(1):32.
Zhao Z, et al. Prediction model and risk scores of ICU admission and mortality in COVID-19. PLoS ONE. 2020;15(7): e0236618.
Bastug A, et al. Clinical and laboratory features of COVID-19: predictors of severe prognosis. Int Immunopharmacol. 2020;88: 106950.
Bellos I, et al. Development of a novel risk score for the prediction of critical illness amongst COVID-19 patients. Int J Clin Pract. 2021;75(4): e13915.
Cheng FY, et al. Using machine learning to predict ICU transfer in hospitalized COVID-19 patients. J Clin Med. 2020;9(6):1668.
Durmus Kocak N, et al. Use of radiology, D-Dimer, and mean platelet volume combination as a prognostic marker in hospitalized coronavirus disease-19 patients. Front Med (Lausanne). 2021;8: 788551.
Geraili Z, et al. Prognostic accuracy of inflammatory markers in predicting risk of ICU admission for COVID-19: application of time-dependent receiver operating characteristic curves. J Int Med Res. 2022;50(6):3000605221102217.
Guner R, et al. Development and validation of nomogram to predict severe illness requiring intensive care follow up in hospitalized COVID-19 cases. BMC Infect Dis. 2021;21(1):1004.
Hachim MY, et al. D-dimer, troponin, and urea level at presentation With COVID-19 can predict ICU admission: a single centered study. Front Med (Lausanne). 2020;7: 585003.
Hashem MK, et al. Prognostic biomarkers in COVID-19 infection: value of anemia, neutrophil-to-lymphocyte ratio, platelet-to-lymphocyte ratio, and D-dimer. Egypt J Bronchol. 2021;15(1):1–9.
Heo J, et al. Prediction of patients requiring intensive care for COVID-19: development and validation of an integer-based score using data from centers for disease control and prevention of South Korea. J Intensive Care. 2021;9(1):16.
Huang HF, et al. Validated tool for early prediction of intensive care unit admission in COVID-19 patients. World J Clin Cases. 2021;9(28):8388–403.
Huespe IA, et al. Multicenter validation of early warning scores for detection of clinical deterioration in COVID-19 hospitalized patients. Med Intensiva. 2021. https://doi.org/10.1016/j.medine.2021.11.023.
Ketenci S, et al. Retrospective analysis of biochemical markers in COVID-19 intensive care unit patients. Egypt J Bronchol. 2022;16(1):27.
Paranjape N, et al. Development and validation of a predictive model for critical illness in adult patients requiring hospitalization for COVID-19. PLoS ONE. 2021;16(3): e0248891.
Shanbehzadeh M, Nopour R, Kazemi-Arpanahi H. Using decision tree algorithms for estimating ICU admission of COVID-19 patients. Inform Med Unlocked. 2022;30: 100919.
Statsenko Y, et al. Prediction of COVID-19 severity using laboratory findings on admission: informative values, thresholds, ML model performance. BMJ Open. 2021;11(2): e044500.
Wira Suastika NK, Suega K. The use of eosinophil count in predicting the need of coronavirus disease 2019 patient for treatment in intensive care unit. Open Access Macedonian J Med Sci. 2021;9:631–5.
Alberdi-Iglesias A, et al. Role of SpO2/FiO2 ratio and ROX index in predicting early invasive mechanical ventilation in COVID-19. A pragmatic, retrospective, multi-center study. Biomedicines. 2021;9(8):1036.
Amezcua-Guerra LM, et al. A simple and readily available inflammation-based risk scoring system on admission predicts the need for mechanical ventilation in patients with COVID-19. Inflamm Res. 2021;70(6):731–42.
Ashkenazi M, et al. A practical clinical score predicting respiratory failure in COVID-19 patients. Isr Med Assoc J. 2022;24(5):327–31.
Garcia-Gordillo JA, et al. COVID-IRS: a novel predictive score for risk of invasive mechanical ventilation in patients with COVID-19. PLoS ONE. 2021;16(4): e0248357.
Kulkarni AR, et al. Deep learning model to predict the need for mechanical ventilation using chest X-ray images in hospitalised patients with COVID-19. BMJ Innov. 2021;7(2):261–70.
Li W, et al. Early predictors for mechanical ventilation in COVID-19 patients. Ther Adv Respir Dis. 2020;14:1753466620963017.
Aguirre-Garcia GM, et al. Stratifying risk outcomes among adult COVID-19 inpatients with high flow oxygen: the R4 score. Pulmonology. 2021. https://doi.org/10.1016/j.pulmoe.2021.10.001.
Alessandri F, et al. Use of CPAP failure score to predict the risk of helmet-CPAP support failure in COVID-19 patients: a retrospective study. J Clin Med. 2022;11(9):2953.
Alvarez-Uria G, et al. Development and validation of the RCOS prognostic index: a bedside multivariable logistic regression model to predict hypoxaemia or death in patients with SARS-CoV-2 infection. Interdiscip Perspect Infect Dis. 2022;2022:2360478.
Aly MM, et al. Can hematological ratios predict outcome of COVID-19 patients? A Multicentric Study J Blood Med. 2021;12:505–15.
Arvind V, et al. Development of a machine learning algorithm to predict intubation among hospitalized patients with COVID-19. J Crit Care. 2021;62:25–30.
Cervantes-Alvarez E, et al. Galectin-3 as a potential prognostic biomarker of severe COVID-19 in SARS-CoV-2 infected patients. Sci Rep. 2022;12(1):1856.
Cruciata A, et al. Risk of seven-day worsening and death: a new clinically derived COVID-19 score. Viruses. 2022;14(3):642.
de Alencar JCG, et al. Lung ultrasound score predicts outcomes in COVID-19 patients admitted to the emergency department. Ann Intensive Care. 2021;11(1):6.
Downing J, et al. Predictors of intubation in COVID-19 patients undergoing awake proning in the emergency department. Am J Emerg Med. 2021;49:276–86.
Faria SP, et al. Forecasting COVID-19 severity by intelligent optical fingerprinting of blood samples. Diagnostics (Basel). 2021;11(8):1309.
Giamarellos-Bourboulis EJ, et al. Development and validation of SCOPE score: a clinical score to predict COVID-19 pneumonia progression to severe respiratory failure. Cell Rep Med. 2022;3(3): 100560.
Gonzalez-Flores J, et al. Usefulness of easy-to-use risk scoring systems rated in the emergency department to predict major adverse outcomes in hospitalized COVID-19 patients. J Clin Med. 2021;10(16):3657.
Gorgojo-Galindo O, et al. Predictive modeling of poor outcome in severe COVID-19: a single-center observational study based on clinical, cytokine and laboratory profiles. J Clin Med. 2021;10(22):5431.
Gresser E, et al. Risk stratification for ECMO requirement in COVID-19 ICU patients using quantitative imaging features in CT scans on admission. Diagnostics (Basel). 2021;11(6):1029.
Gude F, et al. Development and validation of a clinical score to estimate progression to severe or critical state in COVID-19 pneumonia hospitalized patients. Sci Rep. 2020;10(1):19794.
Hao B, et al. Development and validation of predictive models for COVID-19 outcomes in a safety-net hospital population. J Am Med Inform Assoc. 2022;29(7):1253–62.
He F, et al. The development and validation of simplified machine learning algorithms to predict prognosis of hospitalized patients With COVID-19: multicenter, retrospective study. J Med Internet Res. 2022;24(1): e31549.
Hiremath A, et al. Integrated clinical and CT based artificial intelligence nomogram for predicting severity and need for ventilator support in COVID-19 patients: a multi-site study. IEEE J Biomed Health Inform. 2021;25(11):4110–8.
Huang D, et al. Blood urea nitrogen to serum albumin ratio (BAR) predicts critical illness in patients with coronavirus disease 2019 (COVID-19). Int J Gen Med. 2021;14:4711–21.
Jimenez-Solem E, et al. Developing and validating COVID-19 adverse outcome risk prediction models from a bi-national European cohort of 5594 patients. Sci Rep. 2021;11(1):3246.
Kamran F, et al. Early identification of patients admitted to hospital for covid-19 at risk of clinical deterioration: model development and multisite external validation study. BMJ. 2022;376: e068576.
Kim HJ, et al. Validation of machine learning models to predict adverse outcomes in patients with COVID-19: a prospective pilot study. Yonsei Med J. 2022;63(5):422–9.
Knight SR, et al. Prospective validation of the 4C prognostic models for adults hospitalised with COVID-19 using the ISARIC WHO clinical characterisation protocol. Thorax. 2022;77(6):606–15.
Kucuk M, et al. The predictive values of respiratory rate oxygenation index and chest computed tomography severity score for high-flow nasal oxygen failure in critically Ill patients with coronavirus disease-2019. Balkan Med J. 2022;39(2):140–7.
Lombardi Y, et al. External validation of prognostic scores for COVID-19: a multicenter cohort study of patients hospitalized in greater Paris University Hospitals. Intensive Care Med. 2021;47(12):1426–39.
Magunia H, et al. Machine learning identifies ICU outcome predictors in a multicenter COVID-19 cohort. Crit Care. 2021;25(1):295.
Mu E, et al. Augmenting existing deterioration indices with chest radiographs to predict clinical deterioration. PLoS ONE. 2022;17(2): e0263922.
Pournazari P, et al. Cardiac involvement in hospitalized patients with COVID-19 and its incremental value in outcomes prediction. Sci Rep. 2021;11(1):19450.
Shanbhag V, et al. Utility of age-adjusted Charlson comorbidity index as a predictor of need for invasive mechanical ventilation, length of hospital stay, and survival in COVID-19 patients. Indian J Crit Care Med. 2021;25(9):987–91.
Tevald MA, et al. Activity measure for post-acute care “6-Clicks” for the prediction of short-term clinical outcomes in individuals hospitalized With COVID-19: a retrospective cohort study. Arch Phys Med Rehabil. 2021;102(12):2300-2308e3.
Torres-Macho J, et al. Prediction accuracy of serial lung ultrasound in COVID-19 hospitalized patients (Pred-Echovid Study). J Clin Med. 2021;10(21):4818.
Valencia CF, et al. Comparison of ROX and HACOR scales to predict high-flow nasal cannula failure in patients with SARS-CoV-2 pneumonia. Sci Rep. 2021;11(1):22559.
Varghese BA, et al. Predicting clinical outcomes in COVID-19 using radiomics on chest radiographs. Br J Radiol. 2021;94(1126):20210221.
Venturini S, et al. Development and validation of the acute pneumonia early assessment score for safely discharging low-risk SARS-CoV-2-infected patients from the emergency department. J Clin Med. 2022;11(3):881.
Youssef A, et al. Development and validation of early warning score systems for COVID-19 patients. Healthc Technol Lett. 2021;8(5):105–17.
Yu L, et al. Machine learning methods to predict mechanical ventilation and mortality in patients with COVID-19. PLoS ONE. 2021;16(4): e0249285.
Zhou J, et al. Development of a multivariable prediction model for severe COVID-19 disease: a population-based study from Hong Kong. NPJ Digit Med. 2021;4(1):66.
Wynants L, et al. Prediction models for diagnosis and prognosis of covid-19: systematic review and critical appraisal. BMJ. 2020;369: m1328.
Mallett S, et al. Reporting methods in studies developing prognostic models in cancer: a review. BMC Med. 2010;8:20.
Collins GS, et al. Transparent reporting of a multivariable prediction model for individual prognosis or diagnosis (TRIPOD): the TRIPOD Statement. BMC Med. 2015;13:1.
Shipe ME, et al. Developing prediction models for clinical use using logistic regression: an overview. J Thorac Dis. 2019;11(Suppl 4):S574-s584.
Austin PC, Steyerberg EW. Events per variable (EPV) and the relative performance of different strategies for estimating the out-of-sample validity of logistic regression models. Stat Methods Med Res. 2017;26(2):796–808.
Harrell FE Jr, Lee KL, Mark DB. Multivariable prognostic models: issues in developing models, evaluating assumptions and adequacy, and measuring and reducing errors. Stat Med. 1996;15(4):361–87.
Twohig KA, et al. Hospital admission and emergency care attendance risk for SARS-CoV-2 delta (B.1.617.2) compared with alpha (B.1.1.7) variants of concern: a cohort study. Lancet Infect Dis. 2022;22(1):35–42.
Gupta RK, et al. Systematic evaluation and external validation of 22 prognostic models among hospitalised adults with COVID-19: an observational cohort study. Eur Respir J. 2020;56(6):2003498.
Raschke RA, et al. Discriminant accuracy of the SOFA score for determining the probable mortality of patients With COVID-19 pneumonia requiring mechanical ventilation. JAMA. 2021;325(14):1469–70.
Yang X, et al. Clinical course and outcomes of critically ill patients with SARS-CoV-2 pneumonia in Wuhan, China: a single-centered, retrospective, observational study. Lancet Respir Med. 2020;8(5):475–81.
Keller MB, et al. Preintubation sequential organ failure assessment score for predicting COVID-19 mortality: external validation using electronic health record from 86 US healthcare systems to appraise current ventilator triage algorithms. Crit Care Med. 2022;50(7):1051–62.
Funding
Open access funding provided by University of Bern. This project has received funding from: (i) European Union's Horizon 2020 research and innovation programme under grant agreement No. 101017915 and (ii) Swiss National Science Foundation (SNF): #320030_176216.
Author information
Authors and Affiliations
Contributions
All authors listed have made a substantial, direct, and intellectual contribution to the work, as well as approved it for publication.
Corresponding author
Ethics declarations
Conflict of interest
The authors have no disclosures to report.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Buttia, C., Llanaj, E., Raeisi-Dehkordi, H. et al. Prognostic models in COVID-19 infection that predict severity: a systematic review. Eur J Epidemiol 38, 355–372 (2023). https://doi.org/10.1007/s10654-023-00973-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10654-023-00973-x