Abstract
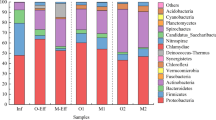
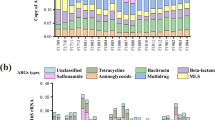
The occurrence of antibiotic-resistant bacteria and antibiotic resistance genes (ARGs) has been intensively investigated for wastewater treatment systems treating single class of antibiotic in recent years. However, the impacts of alternately occurring antibiotics in antibiotic production wastewater on the behavior of ARGs in biological treatment systems were not well understood yet. Herein, techniques including high-capacity quantitative PCR and quantitative PCR (qPCR) were used to investigate the behavior of ARGs in an anaerobic–aerobic full-scale system. The system alternately treated three kinds of antibiotic production wastewater including ribostamycin, spiramycin and paromomycin, which referred to stages 1, 2 and 3. The aminoglycoside ARGs (52.1–79.3%) determined using high-capacity quantitative PCR were the most abundant species in all sludge samples of the three stages. The total relative abundances of macrolide–lincosamide–streptogramin (MLS) resistance genes and aminoglycoside resistance genes measured using qPCR were significantly higher (P < 0.05) in aerobic sludge than in sewage sludge. However, the comparison of ARGs acquired from three alternate stages revealed that MLS genes and the aminoglycoside ARGs did not vary significantly (P > 0.05) in both aerobic and anaerobic sludge samples. In aerobic sludge, one acetyltransferase gene (aacA4) and the other three nucleotidyltransferase genes (aadB, aadA and aadE) exhibited positive correlations with intI1 (r 2 = 0.83–0.94; P < 0.05), implying the significance of horizontal transfer in their proliferation. These results and facts will be helpful to understand the abundance and distribution of ARGs from antibiotic production wastewater treatment systems.





Similar content being viewed by others
References
Aydin, S., Ince, B., & Ince, O. (2015). Development of antibiotic resistance genes in microbial communities during long-term operation of anaerobic reactors in the treatment of pharmaceutical wastewater. Water Research, 83, 337–344.
Ben Said, L., Klibi, N., Lozano, C., Dziri, R., Ben Slama, K., Boudabous, A., et al. (2015). Diversity of enterococcal species and characterization of high-level aminoglycoside resistant enterococci of samples of wastewater and surface water in Tunisia. Science of the Total Environment, 530–531, 11–17.
Chen, J., Ying, G. G., Wei, X. D., Liu, Y. S., Liu, S. S., Hu, L. X., et al. (2016). Removal of antibiotics and antibiotic resistance genes from domestic sewage by constructed wetlands: Effect of flow configuration and plant species. Science of the Total Environment, 571, 974–982.
Chen, J., Yu, Z., Michel, F. C., Wittum, T., & Morrison, M. (2007). Development and application of real-time PCR assays for quantification of erm genes conferring resistance to macrolides–lincosamides–streptogramin B in livestock manure and manure management systems. Applied and Environmental Microbiology, 73(14), 4407–4416.
Devarajan, N., Laffite, A., Graham, N. D., Meijer, M., Prabakar, K., Mubedi, J. I., et al. (2015). Accumulation of clinically relevant antibiotic-resistance genes, bacterial load, and metals in freshwater lake sediments in Central Europe. Environmental Science and Technology, 49(11), 6528–6537.
Fernandez-Martinez, M., Miro, E., Ortega, A., Bou, G., Gonzalez-Lopez, J. J., Oliver, A., et al. (2015). Molecular identification of aminoglycoside-modifying enzymes in clinical isolates of Escherichia coli resistant to amoxicillin/clavulanic acid isolated in Spain. International Journal of Antimicrobial Agents, 46(2), 157–163.
Gillings, M., Boucher, Y., Labbate, M., Holmes, A., Krishnan, S., Holley, M., et al. (2008). The evolution of class 1 integrons and the rise of antibiotic resistance. Journal of Bacteriology, 190(14), 5095–5100.
Harb, M., Wei, C. H., Wang, N., Amy, G., & Hong, P. Y. (2016). Organic micropollutants in aerobic and anaerobic membrane bioreactors: Changes in microbial communities and gene expression. Bioresource Technology, 218, 882–891.
Koike, S., Aminov, R. I., Yannarell, A. C., Gans, H. D., Krapac, I. G., Chee-Sanford, J. C., et al. (2010). Molecular ecology of macrolide–lincosamide–streptogramin B methylases in waste lagoons and subsurface waters associated with swine production. Microbial Ecology, 59(3), 487–498.
Li, D., Yu, T., Zhang, Y., Yang, M., Li, Z., Liu, M. M., et al. (2010). Antibiotic resistance characteristics of environmental bacteria from an oxytetracycline production wastewater treatment plant and the receiving river. Applied and Environmental Microbiology, 76(11), 3444–3451.
Liu, M., Ding, R., Zhang, Y., Gao, Y., Tian, Z., Zhang, T., et al. (2014). Abundance and distribution of Macrolide–Lincosamide–Streptogramin resistance genes in an anaerobic–aerobic system treating spiramycin production wastewater. Water Research, 63, 33–41.
Liu, M., Zhang, Y., Yang, M., Tian, Z., Ren, L., & Zhang, S. (2012). Abundance and distribution of tetracycline resistance genes and mobile elements in an oxytetracycline production wastewater treatment system. Environmental Science and Technology, 46(14), 7551–7557.
Luby, E. M., Moorman, T. B., & Soupir, M. L. (2016). Fate and transport of tylosin-resistant bacteria and macrolide resistance genes in artificially drained agricultural fields receiving swine manure. Science of the Total Environment, 550, 1126–1133.
Ma, Y., Wilson, C. A., Novak, J. T., Riffat, R., Aynur, S., Murthy, S., et al. (2011). Effect of various sludge digestion conditions on sulfonamide, macrolide, and tetracycline resistance genes and class I integrons. Environmental Science and Technology, 45(18), 7855–7861.
Marti, E., Jofre, J., & Balcazar, J. L. (2013). Prevalence of antibiotic resistance genes and bacterial community composition in a river influenced by a wastewater treatment plant. PLoS ONE, 8(10), 1–8.
Mingeot-Leclercq, M. P., Glupczynski, Y., & Tulkens, P. M. (1999). Aminoglycosides: Activity and resistance. Antimicrobial Agents and Chemotherapy, 43(4), 727–737.
Ramirez, M. S., & Tolmasky, M. E. (2010). Aminoglycoside modifying enzymes. Drug Resistance Updates: Reviews and Commentaries in Antimicrobial and Anticancer Chemotherapy, 13(6), 151–171.
Roberts, M. C., Sutcliffe, J., Courvalin, P., Jensen, L. B., Rood, J., & Seppala, H. (1999). Nomenclature for macrolide and macrolide–lincosamide–streptogramin B resistance determinants. Antimicrobial Agents and Chemotherapy, 43(12), 2823–2830.
Schmittgen, T. D., & Livak, K. J. (2008). Analyzing real-time PCR data by the comparative CT method. Nature Protocols, 3(6), 1101–1108.
Schönfeld, W., & Kirst, H. A. (2002). Macrolide antibiotics (pp. 281–305). Berlin: Springer Basel AG.
Sidrach-Cardona, R., Hijosa-Valsero, M., Marti, E., Balcazar, J. L., & Becares, E. (2014). Prevalence of antibiotic-resistant fecal bacteria in a river impacted by both an antibiotic production plant and urban treated discharges. The Science of the Total Environment, 488–489, 220–227.
Su, J. Q., Wei, B., Ou-Yang, W. Y., Huang, F. Y., Zhao, Y., Xu, H. J., et al. (2015). Antibiotic resistome and its association with bacterial communities during sewage sludge composting. Environmental Science and Technology, 49(12), 7356–7363.
Tian, Z., Zhang, Y., Yu, B., & Yang, M. (2016). Changes of resistome, mobilome and potential hosts of antibiotic resistance genes during the transformation of anaerobic digestion from mesophilic to thermophilic. Water Research, 98, 261–269.
Topp, E., Renaud, J., Sumarah, M., & Sabourin, L. (2016). Reduced persistence of the macrolide antibiotics erythromycin, clarithromycin and azithromycin in agricultural soil following several years of exposure in the field. Science of the Total Environment, 562, 136–144.
Vakulenko, S. B., & Mobashery, S. (2003). Versatility of aminoglycosides and prospects for their future. Clinical Microbiology Reviews, 16(3), 430–450.
Wang, J. L., Mao, D. Q., Mu, Q. H., & Luo, Y. (2015). Fate and proliferation of typical antibiotic resistance genes in five full-scale pharmaceutical wastewater treatment plants. Science of the Total Environment, 526, 366–373.
Wang, F. H., Qiao, M., Su, J. Q., Chen, Z., Zhou, X., & Zhu, Y. G. (2014). High throughput profiling of antibiotic resistance genes in urban park soils with reclaimed water irrigation. Environmental Science and Technology, 48(16), 9079–9085.
Wei, F. S. (2002). Monitoring and analysis methods of water and wastewater, 4th ed. (pp. 210–213). Beijing: China Environmental Science Press.
Yu, X., Zuo, J., Li, R., Gan, L., Li, Z., & Zhang, F. (2014). A combined evaluation of the characteristics and acute toxicity of antibiotic wastewater. Ecotoxicology and Environmental Safety, 106, 40–45.
Zhai, W. C., Yang, F. X., Mao, D. Q., & Luo, Y. (2016). Fate and removal of various antibiotic resistance genes in typical pharmaceutical wastewater treatment systems. Environmental Science and Pollution Research, 23(12), 12030–12038.
Zhang, Y., Xie, J., Liu, M., Tian, Z., He, Z., van Nostrand, J. D., et al. (2013). Microbial community functional structure in response to antibiotics in pharmaceutical wastewater treatment systems. Water Research, 47(16), 6298–6308.
Zhang, H., Zhang, Y., Yang, M., & Liu, M. (2015). Evaluation of residual antibacterial potency in antibiotic production wastewater using a real-time quantitative method. Environmental Science Processes and Impacts, 17(11), 1923–1929.
Zhu, Y. G., Johnson, T. A., Su, J. Q., Qiao, M., Guo, G. X., Stedtfeld, R. D., et al. (2013). Diverse and abundant antibiotic resistance genes in Chinese swine farms. Proceedings of the National Academy of Sciences of the United States of America, 110(9), 3435–3440.
Acknowledgements
This project is supported by National Natural Scientific Foundation of China (No. 21437005) and special fund of State Key Joint Laboratory of Environmental Simulation and Pollution Control (15L03ESPC).
Author information
Authors and Affiliations
Corresponding authors
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Tang, M., Dou, X., Wang, C. et al. Abundance and distribution of antibiotic resistance genes in a full-scale anaerobic–aerobic system alternately treating ribostamycin, spiramycin and paromomycin production wastewater. Environ Geochem Health 39, 1595–1605 (2017). https://doi.org/10.1007/s10653-017-9987-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10653-017-9987-5