Abstract
Municipal wastewater treatment plants (mWWTPs), considered reservoirs of antibiotic resistance genes (ARGs), are selected to compare the contributions of technology and process to ARG removal. Fifteen ARGs (tetA, tetB, tetC, tetE, tetG, tetL, tetM, tetO, tetQ, tetS, tetX, MOX, CIT, EBC, and FOX) and two integron genes (intI1, intI2) were tracked and detected in wastewater samples from a large-scale mWWTP with four parallel processes, including three biological technologies of AAO (anaerobic-anoxic-oxic), AB (adsorption-biodegradation), and UNITANK, two different disinfection technologies, and two primary sedimentation steps. The results showed that ARGs were widely detected, among which tetA and tetM had the highest detection rate at 100%. AAO was the most effective process in removing ARGs, followed by the AB and UNITANK processes, where the separation step was critical: 37.5% AmpC β-lactamase genes were reduced by the secondary clarifier. UV disinfection was more efficient than chlorination disinfection by 47.0% in ARG removal. Both disinfection and primary sedimentation processes could effectively remove integrons, and the swirling flow grit chamber was a more effective primary settling facility in total ARG removal than the aerated grit chamber. The tet genes and AmpC β-lactamase genes were significantly correlated with the water quality indexes of BOD5, CODCr, SS, TP, TOC, pH and NH4+-N (p < 0.05). In addition, the correlation between efflux pump genes and AmpC β-lactamase genes was strongly significant (r2 = 0.717, p < 0.01). This study provides a more powerful guide for selecting and designing treatment processes in mWWTPs with additional consideration of ARG removal.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Tetracycline and β-lactam are two types of antibiotics predominately used in human welfare and livestock as prophylaxis and therapy, with residual antibiotics contaminating soil and water (Widyasari-Mehta et al. 2016; Zhang et al. 2015). Due to repeated exposure to antibiotics, environmental microorganisms develop resistance against antibiotics and survive, which exacerbates the problem of the emergence of antibiotic-resistant bacteria (ARB) and ARGs (Kim et al. 2018). To date, at least forty different tetracycline resistance genes have been detected in various environments, such as aquatic environments (Li et al. 2020), wetlands (Li et al. 2019), mangrove ecosystems (Liu et al. 2020), WWTPs (Chen, Zhang 2013a; b), hospital wastewaters (He et al. 2020), and livestock farms (Duan et al. 2019). Six urban lakes in Wuhan detected antibiotic efflux pumps and ribosomal protection protein genes (tetA, tetB, tetC, tetG and tetM, tetQ) (Yang et al. 2017). Microorganisms may use these pollutants in ecosystems to assimilate and transform (Gu. 2019). Different types of AmpC β-lactamase genes have also been found in many gram-negative bacteria, including Acinetobacter (Wang et al. 2008), Aeromonas caviae (Ye et al. 2010), Proteus mirabilis (Ibrahimagić et al. 2015), Providencia spp. (Mahrouki et al. 2015), Escherichia coli (Bajaj et al. 2015), and Klebsiella pneumoniae (Venieri et al. 2017). ARGs can disseminate within or among species via heredity and horizontal gene transfer (HGT), which means that ARGs can be transferred from one bacterial strain to another (Zhang et al. 2018). The frequency and rapid spread of acquired plasmid-mediated AmpC β-lactamases are increasing among Enterobacteriaceae worldwide, and infections caused by resistant Enterobacteriaceae due to AmpC have increased the morbidity and mortality rate compared with those caused by susceptible Enterobacteriaceae (Etemadi et al. 2020). Higher concentrations of ARGs are easy to transfer to the human body, especially the human intestinal tract. In addition, mobile genetic elements such as conjugative plasmids frequently carry genes other than ARGs that contribute to microbial fitness (McInnes et al. 2020). When these pollutants are bio-enriched to a certain concentration, they will have a toxic effect on the entire ecology and affect the balance of the ecosystem. The induced ARGs, as emerging environmental contaminants, cause global threats to human society (Organization 2014).
Untreated wastewater usually contains a large number of pathogens. Souissi et al. (2018) isolated and identified Leuconostoc spp., Chryseomona luteola, and Staphylococcus xylosus from WWTP wastewater. Medema et al. (2020) reported that SARS-CoV-2 viral RNA was first detected in the influent of WWTPs in the Netherlands. Fecal streptococci and pathogen Staphylococci groups contained serious β-lactams or aminoglycosides and acquired resistance (Souissi et al. 2018). New research has shown that the quantity of ARGs in effluents is related to the size of WWTPs, and the abundance of ARGs is highest in small WWTPs (Harnisz et al. 2020). Conventional WWTPs generally fail to effectively reduce ARGs, especially extracellular ARGs (Li et al. 2019). The AAO process, as a widespread biological nutrient removal configuration, can achieve simultaneous removal of organics (Huang et al. 2020). However, it was further revealed that tetA, tetB, tetC, tetE, tetM, tetO, tetS, and tetX were common and abundant in all reactors of the improved AAO in WWTPs (Huang et al. 2015). The AB process, also called the two-stage activated sludge process, has a high removal rate of organic substrates. The UNITANK process is an improvement of the sequencing batch reactor (SBR), in which wastewater is added to a single batch reactor and returning sludge is eliminated (Bashiri et al. 2018). However, it is prone to sludge deposition (Liu et al. 2018), which easily causes ARGs from wastewater to be transferred into sludge in both the AB and UNITANK processes. Higher amounts of some ARGs were detected during the pretreatment process, including influent and primary sedimentation in AAO, but decreased gradually via sequential treatment processes (Lee et al. 2017). Although both UV and chlorination disinfection could affect the reduction of ARGs, chlorination might augment the risk of ARG transfer in wastewater containing NH3-N (Sharma et al. 2016). The disadvantage of the AB process is that the A stage is prone to produce more sludge. The genes tetA, tetM, tetW, and tetX were a large proportion of the sludge samples. (Xu et al. 2020; Zhou et al. 2019; Zhang et al. 2019). However, in the presence of trace tetracycline, the relative abundances of efflux pumps, such as tetA and tetG, tended to remarkably increase in the UNITANK process (Liu et al. 2019). Overall, the conventional and advanced processes resulted in 0.03–2.40 log reductions for most ARG subtypes, such as intI1, qepA, strA, and strB, but neither treatment method affected the reduction of tetM (Hu et al. 2019). Traditionally, a combination of physical, chemical, and biological processes has been widely used in wastewater treatment plants to remove pollutants (Li et al. 2017), such as BOD5, CODCr, SS, TP, TN, etc. and ensure that the residual pollutants in the effluent conform to the legal requirements (Everage et al. 2014; Jaranowska et al. 2013), so the reduction of ARGs was not considered during the treatment process (De Sotto et al. 2016). WWTPs have been considered a reservoir of ARGs because a high abundance and diversity of ARGs were detected in WWTPs, such as quinolone resistance genes qnrS, qnrB, and acc(6’)-Ib-cr, AmpC genes, integrons intI1 and intI2 (Su et al. 2014; Kumar et al. 2020), tet genes tetM tetC tetK and tetA/P (Laht et al. 2014; Zhang, Zhang 2011), which might be released into the drinking water from the effluent and pose a potential risk to human health. Excessive residues of ARGs have caused serious threats to human and animal health due to the spread of antibiotics. To make the distribution of ARGs clear and raise the management level of ARGs, it is important to study the relationship between the present wastewater treatment process and the removal of ARGs.
In this study, a large-scale mWWTP in Guangzhou, which contains four different treatment processes, was selected to investigate the relationship between the treatment process and elimination of ARGs. Eleven tetracycline resistance genes (six efflux pump genes: tetA, tetB, tetC, tetE, tetG, tetL, four ribosomal protection genes: tetM, tetO, tetQ, tetS, one enzymatic modification gene: tetX), four family-specific AmpC β-lactamase genes (MOX, CIT, EBC, FOX), and two integron genes (intI1, intI2) were detected by using polymerase chain reaction (PCR) and real-time qualitative polymerase chain reaction (qPCR). According to the same properties of the influent and the same operation level in the plant, the results provide strong evidence for selecting the key process step and disinfection method to control ARG discharge, which will benefit the reduction of health and ecosystem risk.
Materials and methods
Municipal wastewater treatment plants
A specialized large-scale municipal wastewater treatment plant in Guangzhou was selected to collect samples. This plant treats the wastewater for an equivalent population of 2.96 million and has a maximum capacity of 560,000 m3/d, occupying an area of 390,000 m2 and receiving the wastewater from the central city area of 228 km2 belonging to the North Waterway of the Pearl River. It is constructed in four phases, and the basic information is shown in Table S1. The wastewater from the same conditioner with the same properties is pumped into four phases of proceedings, and effluent treated by different processes and disinfection methods is discharged through the same draining exit.
Sample collection and pretreatment
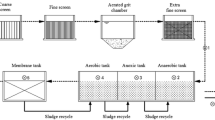
Twenty-one samples of water were collected from the influent, effluent, and each treatment unit of the mWWTP, as shown in Fig. 1. At each sampling location, a 2000 mL water sample was transferred to two sterile 1000 mL bottles and stored at 4 °C before pretreatment within 24 h. Chemical parameters of influent and effluent samples from four phases were detected and recorded (Table S2).
Treatment process flow charts and sampling locations of the four phases with different process. A The influent of the grid screen as the influent of the main process. (B1-2) Two different primary sedimentation tanks, Phase I(AB process) and Phase II(UNITANK process) share a swirling flow frit chamber, and Phase III(Modified AAO process) and Phase IV(Modified AAO process) share an aerated grit chamber. Phase I: (C1) the mixture in the stage B aerobic tank, (C2) the effluent of the stage B clarifier. Phase II: (D1) the mixture in the A tank, (D2) the mixture in the B tank, (D3) the effluent of the UNITANK. Phase I-II, (R1) the disinfection effluent of Phase I and II. Phase III: (E1) the mixture in the pre-anoxic tank, (E2) the mixture in the anaerobic tank, (E3) the mixture in the anoxic tank, (E4) the mixture in the aerobic tank, (E5) the secondary clarifier effluent, and (E6) the disinfection by chlorination. Phase IV (Modified AAO process): (F1-5) same as Phase III and (F6) the disinfection by UV
DNA extraction
One thousand milliliters of water samples were filtered through 50 mm cellulose ester membranes with pore sizes of 0.22 μm. The filters were stored at −20 °C to protect DNA before extraction. Total DNA was extracted using the E.Z.N.A. TM Water DNA Kit (Omerga Bio-tek, USA) according to the manufacturer’s protocol. The quality and concentration of the purified DNA were determined by spectrophotometer analysis (NanoDrop 1000, Thermo Scientific, USA) (He et al. 2017) and quantified by 1.5% agarose gel electrophoresis. The total DNA samples were stored at −20 °C pending further analysis.
Qualitative PCR
Fifteen ARGs (tetA, tetB, tetC, tetE, tetG, tetL, tetM, tetO, tetQ, tetS, tetX, MOX, CIT, EBC, FOX) and two integron genes (intI1, intI2) in all water samples were investigated by using conventional PCR with the primers listed in Table S3. The reaction volume of PCR was 25 μL consisting of 2.5 μL of 10 × PCR Buffer (Mg2+ Plus, TaKaRa, Japan), 2.0 μL of dNTP Mixture (each 2.5 mM, TaKaRa, Japan), 1.0 μL of each forward and reverse primer (10 μM), 0.125 μL of Taq polymerase (5 U/μL, TaKaRa, Japan), and 1 μL of DNA template, and 17.375 μL of ddH2O. The PCR program was performed on a thermal cycler (Eppendorf, Germany) as follows: initial denaturation at 94 °C for 5 min, followed by 30 cycles of 94 °C for 30 s, annealing at 55 °C for 30 s, 72 °C for 60 s, and a final extension of 72 °C for 7 min. PCR products were analyzed by electrophoresis on a 1.5% (w/v) agarose gel with ethidium bromide in 0.5× TBE buffer at 100 V for 40 min and visualized under UV transillumination. Ultrapure water was used as the negative control.
ARG quantitative analysis
The quantities of target ARGs, integrons, and 16S rRNA were detected by qPCR using the SYBR® Green approach and specific primers. The 20 μL qPCR reaction mixtures consisted of 7 μL of ddH2O, 10 μL of 2 × iTaqTM universal SYBR® Green supermix (Bio-Rad, USA), 2 μL of forward and reverse primers (10 μM of each type), and 1 μL of template DNA (Table S4). Amplification was conducted with CFX (Bio-Rad, USA) as follows: initial denaturation at 95 °C for 3 min, 40 cycles at 95 °C for 10 s, and 30 s with the plate read at the annealing temperature of 58 °C. Immediately after the qPCR assay, melting curve analyses were performed by increasing the temperature from 65 °C to 95 °C (0.5 °C/5 s) with continuous fluorescence recording according to Huang et al. (2017). Sterile ultrapure water was used as the negative control, and 16S rRNA was determined for each sample as the reference gene. Three independent samples were analyzed at each site, and each sample was quantified in triplicate to ensure reproducibility. The qPCR efficiency of each gene ranged from 90 to 110%, with R2 values greater than 0.990 for all calibration curves.
Statistical analysis
Basic calculations were performed using Microsoft Excel 2016. Correlations between tet genes and AmpC β-lactamase genes were analyzed by SPSS 24.0 statistical software. A variable was considered statistically significant if p < 0.05 or p < 0.01. Data features were analyzed by Origin Pro 8.1 (OriginLab Co., MA, USA).
Results and discussion
Occurrence of ARGs in mWWTP from influent to effluent
For the occurrence of the target ARGs, the detection frequency was 100% in the influent. The genes tetA, tetC, tetM and tetO showed strong persistence throughout the wastewater treatment plant, tetA and tetM were detected in all treatment units (100%), and the detection rate of tetO and tetC in water samples was 90.47% (19/21) (Table 1). However, the gene diversity in the samples was reduced during the treatment process. The quantity of gene types identified in the effluent of phase I-II, phase III, and phase IV were 9, 5, and 3, respectively. A total of 78.57% of the detected ARG types disappeared with the treatment of the whole process used in phase IV, which was the most efficient process for ARG type removal among the four parallel processes used in the mWWTP. Similar results were found that the detection frequencies of tetA, tetM, and tetO were higher than those of other tet genes in WWTPs (Storteboom et al. 2007), and the detection frequency of tetM was 100% (Zhang et al. 2017). The efflux genes (tetA, tetB, tetC, and tetE) and ribosomal protection protein genes (tetM and tetO) are frequently found in various environmental matrices, such as livestock farms (Duan et al. 2019), WWTPs (Xu et al. 2017) and ground water (Wu et al. 2020). The tetracycline resistance genes tetA, tetB, tetC, tetE, tetM, and tetO have a broad host range and are carried by several environmental matrices. Therefore, the strong persistence of these ARGs was due to their broad host range. The ARGs in wastewater were reduced, primarily based on the decrease in total biomass and selective removal of ARGs from bacterial cells (Bengtsson-Palme et al. 2016). Most of the available studies have concluded that WWTPs reduce the absolute numbers of both ARGs and total bacteria in wastewater (Pallares-Vega et al. 2019).
Abundance of ARGs in mWWTP
PCR-detectable tet genes and AmpC β-lactamase genes in each sample were normalized to the 16S rRNA as the relative abundance of genes. The results are shown in Fig. S1. The proportional results indicated that most relative abundances of tet genes were higher than AmpC β-lactamase genes, except for tetB and tetL. In all influent samples, the relative abundance of all resistance genes ranged from 10−3 to 10−5, in which tetX was the highest. However, the residual ARGs were different depending on the process. In Phase I-II, tetB, tetG, tetX, CIT, and FOX were undetected in the effluent, showing that these ARGs can be completely eliminated by the AAO process. For the Phase III effluent, most ARGs could be completely removed except tetA, tetE, tetM, tetO and tetS, while only tetA, tetM and tetS were detected in the effluent from Phase IV. Moreover, the average abundance of ribosomal protection protein genes was higher than that of both efflux pump genes and enzyme-modified genes, which was similar to Cheng et al. (2013). According to published studies, tetracycline resistance genes were found to be more prevalent in bacterial populations than AmpC β-lactamase genes because tetracycline resistance genes could be detected in both gram-positive and gram-negative bacteria (Chopra, Roberts 2001), while AmpC β-lactamase genes were only found in gram-negative bacteria (Jacoby. 2009). Moreover, tetracycline resistance genes could be found on bacterial chromosomes (e.g., tetQ), plasmids (e.g., tetC, tetE, tetK), and transposones (e.g., tetB, tetC, tetE, tetK) (Pazda et al. 2019). These tet genes with high abundance were always carried by mobile elements and could be transferred between bacteria in the environment (Agerso et al. 2007; Huang et al. 2015; Ding et al. 2020).
Comparison of ARG elimination efficiency by processes among AB, UNITANK, and Improved AAO
By comparing phase I-II and phase III with different treatment processes (AB process, UNITANK process, improved AAO process), the gene removal efficiency was higher in phase III than in phase I-II (Fig. 2), e.g., the removal efficiency of tetO in phase I-II and phase III was 19.52% and 72.24%, respectively. However, it was intriguing that the relative abundance of genes in the clarifier of phase II was higher than that in the influent, such as tetA, which increased from 7.32 × 10−3 to 1.23 × 10−2, with a growth rate of 68.17%. This result was similar to the findings that the reduction magnitudes of tetO, tetW, and tetQ in AAO were 2.31 log, 2.13 log, and 2.50 log, respectively (Cheng et al. 2013). The removal rate observed for the tetM gene was an average log reduction of 2.53 ± 0.68 (Pallares-Vega et al. 2019). The tet genes include three types of resistance mechanisms: efflux pump mechanisms (genes encoding energy-dependent efflux proteins), target modification mechanisms (genes encoding ribosomal protection proteins, RPPs), and inactivating enzymes (Chen, Zhang 2013a; b; Huang et al. 2015; Pazda et al. 2019). According to the above results, the efflux pump genes of tetA and tetC and the ribosomal protection protein genes of tetM and tetO were difficult to remove, which may be due to the resistance mechanism (efflux pump mechanism and target modification mechanism). Zhang et al. (2018) also proved that the AAO process could reduce ARGs regardless of the relative abundance or absolute gene copies. This might be due to the proliferation of ARGs after conventional biological treatment processes, which had an influence on microbial growth (Wang et al. 2015). In addition, the plant containing individual process for sludge discharge such as AAO may tolerate to the complicated wastewater. The highest bacterial diversity was achieved in modified AAO process (Yan et al. 2019), which may imply the advantage in ARG removal. The tetracycline resistant bacteria (TRB) declined in the final effluent samples compared to the influent samples (Huang et al. 2015), which may be one of the reasons for the removal of ARGs. The results indicated that the removal efficiency of ARGs in the superior treatment process was better than that in the conventional treatment process, while the effect of the process was improved in the pattern of AAO process > AB process > UNITANK process.
AAO treatment also played an important role in reducing the relative abundance of resistance genes. As shown in Fig. 3, in the aerobic phase, the relative abundance of the four ARGs was effectively reduced, where the removal rates of the efflux pump of resistance genes, ribosome protection genes, enzyme modification genes, and AmpC β-lactamase genes were 29.61%, 63.66%, 67.46%, and 49.1%, respectively. In the anaerobic stage, the removal rate of the ARGs was also high, and the removal rate of tetX was as high as 73.07%. In the preanoxic phase, most ARGs showed growth, in which the AmpC β-lactamase genes were increased by 1.23 times and the efflux pump genes were increased by 32.71%. In the anoxic phase, almost all types of ARGs had increased, with tetX increasing the most, reaching 22.74%, which meant that different ARGs may be attributed to dissolved oxygen or other nutrients. The removal of ARGs may also have a certain relationship with the difference of bacterial species because the oxygen content of each tanks is different. Therefore, in the AAO process stage, the ARG removal capability followed the order aerobic > anaerobic > preanoxic > anoxic.
In addition, the removal of the ARGs in the secondary clarifier also played a crucial role, in which the removal rate of tetX could be as high as 100%, and the removal rate of AmpC β-lactamase genes was also 37.5%. It was found that the removal of the tetracycline resistance gene was more advantageous under aerobic conditions (Su et al. 2019). The ARGs were lower in aerobic tanks and higher in anaerobic tanks (Tao et al. 2014). The results shown in Fig. 3 suggest that biological treatments could more effectively reduce the abundance of tet genes and AmpC β-lactamase genes, which might be related to the high efficiency of WWTPs in reducing the bacterial population (Su et al. 2014). AmpC β-lactamase plays an important role in hydrolyzing all β-lactam antibiotics and contains two types of resistance mechanisms, i.e., chromosomal mediated and plasmid mediated (Mohamudha et al. 2012; Korzeniewska, Harnisz 2013), except cefepime and carbapenems (Maravić et al. 2013; Ebomah and Okoh. 2020). The genes encoding plasmid-mediated AmpC β-lactamase are harbored by mobile elements that could confer transmissible resistance to environmental bacteria and pathogens, which may accelerate cephalosporin resistance dissemination in the environment (Liu et al. 2015; Pazda et al. 2019). Therefore, the detection rates of MOX and EBC were very high (85.71%). In this study, tet genes and AmpC β-lactamase genes were widely found in mWWTP, which indicates a potential health risk in urban areas.
Comparison of ARG elimination efficiency by UV and chlorination disinfection
The relative abundances of ARGs in the effluent in phase III (improved AAO with sodium hypochlorite) and phase IV (improved AAO with ultraviolet disinfection) with the same superior treatment process were different because of different disinfection methods. The removal efficiency for each gene was determined by comparisons of relative abundance between them, which showed that UV disinfection was better than chlorination (Fig. 4). After two different disinfection, the AmpC β-lactamase resistance genes and inhibitory enzyme activity genes were completely removed. However, UV disinfection had removal rates of 99.99% and 97.23% for intI1 and intI2, respectively, and removal rates for intI2 and intI1 for chlorination were only 85.62% and 51.61%, respectively. Among them, UV had a removal rate of 100% for tetG, tetX, MOX, and EBC. Zhou et al. (2020) also found that UV radiation showed significant removal efficiency on ARGs. In phase III, the detection rate of the effluent resistance genes was 43.75% (7/16), while the detection rate of the effluent resistance genes in UV disinfection was 31.25% (5/16). The disinfection may adjust the relative abundance of the microbial genus to the ARGs, but low doses of chlorine stimulated HGT (Wang et al. 2020). UV disinfection may cause damage to ARGs due to direct absorption of ultraviolet light by DNA. UV disinfection can destroy the resistance genes in microorganisms, greatly reducing the spread of genes during horizontal transformation. Mckinney and Pruden (2012) indicated that UV disinfection had the potential to impact ARG damage because DNA absorbs ultraviolet radiation directly. Guo et al. (2014) found that UV could reduce erythromycin resistance gene and tetracycline resistance gene concentrations. In addition, UV intensity and species also had a close relationship with UV disinfection efficiency (Qin et al. 2020). However, Zhang et al. (2015) suggested that the removal efficiency of ARGs by chlorination was better than that with UV. This suggested that sodium hypochlorite might be used for a few stress-tolerant bacteria. A previous study supported this hypothesis that chlorination enriched ARGs and changed the microbial community structure (Shi et al. 2013).
Comparison of ARG removal by different primary sedimentation processes
The primary sedimentation tank is an important treatment for mWWTPs, which mainly removes suspended solids from wastewater. The AB process and UNITANK process share the swirling flow grit chamber, while another two different improved AAO processes share the aerated grit chamber. According to Fig. 4, the proportion of intI1 in the influent reached 54.4%, which was reported to be associated with multiantibiotic resistance (MAR) in various environments (Brooks et al. 2014). Integrons (intI1, intI2) are very important to the migration of ARGs (Zhou et al. 2020); therefore, the removal of integrons is beneficial to reduce the spread of ARGs in the environment. The aerated grit chamber could remove almost all intI1 of the influent, which was obviously more effective than that of the swirling flow grit chamber. However, for intI2, the removal rate by the swirling flow grit chamber was 69.99%, but intI2 in the effluent from the aerated grit chamber was 3 times higher than that in the influent. Jang et al. (2020) also indicated that most target ARGs (tetG, tetH, tetM, tetQ, tetX) showed a significant positive correlation with intI1. From the analysis of the quantity of ARGs and the total abundance, the swirling flow grit chamber was more advantageous than the aerated sand because the CIT was completely removed, and the removal rate of the ribosome protection genes reached 73.88%, which was 54.0% higher than that of the aerated grit chamber. In addition, the gene removal rates of inhibitory enzyme activity, AmpC β-lactamase, and integron reached 67.26%, 72.48%, and 83.29%, respectively. Total microorganisms, integrons, and organic matter removed in wastewater are conducive to the reduction of ARGs (Riquelme Breazeal et al. 2013). The swirling flow grit chamber could remove the ARGs more effectively than the aerated grit chamber, which might be due to the advantages of the low change in the flow rate and high efficiency of the kinetic energy. However, it is still unclear how the abundance and diversity of ARGs are affected by common wastewater treatment processes, especially in mWWTPs, so further investigations should be performed to better address this question.
Correlation of ARGs in mWWTP
The tet genes and the AmpC β-lactamase genes were significantly correlated with BOD5, CODCr, SS, TP, TOC, pH and NH4+-N (p < 0.05), of which tetO and all water quality parameters except pH were significantly correlated (p < 0.01) (Table 2). However, the efflux pump genes tetA, tetB, tetE, and tetL, ribosome protection gene tetS, and AmpC β-lactamase genes MOX and EBC were not significantly correlated with all water quality parameters (p > 0.05). This indicated that some kinds of ARGs in the wastewater were significantly correlated with some physical parameters. Yuan et al. (2018) also proved that most ARGs, including intI1 and tetA, positively correlated with wastewater nutrient (COD, NH3-N, TN and TP) concentrations, and the relative abundance of some ARGs decreased as the quality of wastewater improved. Therefore, for mWWTPs, the removal rate of pollutants in sewage was an important factor affecting the change rate of ARGs.
There was a significant correlation between tet genes and AmpC β-lactamase genes (p < 0.05), in which tetC, tetE, tetM, tetO, tetX, tetO, and CIT were strongly and significantly correlated with FOX (p < 0.01), and tetE, tetL, tetM, and CIT had significant correlations with each other (p < 0.01) (Table 2), which was due to the large number of multidrug resistant bacteria carrying multiple resistance genes in mWWTP. Similarly, Huang et al. (2015) also found a strong significant correlation between tetM and tetO, as well as tetE and tetX (p < 0.01), in an improved AAO process. The total quantities of efflux pump genes, ribosomal protection protein genes, enzymatic modification genes, and AmpC β-actamase genes were significantly correlated, as shown in Table S5. The correlation between the quantity of efflux pump genes was found to be strongly significant with AmpC β-lactamase genes (r2 = 0.717, p < 0.01) and enzyme-modified genes (r2 = 0.523, p < 0.01). It was reported that there was a strong correlation among the total quantity of tet genes (Huang et al. 2015). The correlation between ARGs was affected by not only their relative antibiotics (Cheng et al. 2013; Huang et al. 2015) but also the function of co-selection and cross-selection on resistance from antibiotics and heavy metals (Mckinney et al. 2010; He et al. 2017). Furthermore, water parameters, such as COD, DO, pH, and temperature, have been found to be related to ARGs, and correlations between the removal efficiency of ARGs and the removal efficiency of CODCr, BOD5, nitrogen, and biomass were observed (Nõlvak et al. 2013; Yuan et al. 2016).
Conclusions
Overall treatment processes were carried out in four different phases to evaluate the tet genes and AmpC β-lactamase genes, especially in the effluent, which presented a lower abundance of ARGs. By comparing different processes, the removal efficiency of genes was most significantly improved by the AAO process, followed by the AB process and UNITANK process, while the aerobic tank played an important role in modifying the AAO of ARG reduction. The swirling flow grit chamber was more significant in ARG removal. In addition, ultraviolet disinfection was better than chlorination. The quantity between efflux pump genes and AmpC β-lactamase genes showed a strongly significant correlation (r2 = 0.717, p < 0.01).
References
Agerso Y, Bruun MS, Dalsgaard I, Larsen JL (2007) The tetracycline resistance gene tet(E) is frequently occurring and present on large horizontally transferable plasmids in Aeromonas spp. from fish farms. Aquaculture 266(1–4):47–52. https://doi.org/10.1016/j.aquaculture.2007.01.012
Bajaj P, Singh NS, Kanaujia PK, Virdi JS (2015) Distribution and molecular characterization of genes encoding ctx-m and AmpC β-lactamases in Escherichia coli isolated from an Indian urban aquatic environment. Sci Total Environ 505(86):350. https://doi.org/10.1016/j.scitotenv.2014.09.084
Bashiri B, Fallah N, Bonakdarpour B, Elyasi S (2018) The development of aerobic granules from slaughterhouse wastewater in treating real dyeing wastewater by Sequencing Batch Reactor (SBR). J Environ Chem Eng 6(4):5536–5543. https://doi.org/10.1016/j.jece.2018.05.020
Bengtsson-Palme J, Hammarén R, Pal C, Östman M, Björlenius B, Flach C, Fick J, Kristiansson E, Tysklind M, Larsson DGJ (2016) Elucidating selection processes for antibiotic resistance in sewage treatment plants using metagenomics. Sci Total Environ 572:697–712. https://doi.org/10.1016/j.scitotenv.2016.06.228
Brooks JP, Adeli A, McLaughlin MR (2014) Microbial eclology, bacterial pathogens, and antibiotic resistant genes in swine manure wastewater as influenced by three swine management systems. ScienceDirect 57:96–103. https://doi.org/10.1016/j.watres.2014.03.017
Chen H, Zhang MM (2013a) Effects of advanced treatment systems on the removal of antibiotic resistance genes in wastewater treatment plants from Hangzhou, China. Environ Sci Technol 47(15):8157–8163. https://doi.org/10.1021/es401091y
Chen H, Zhang MM (2013b) Occurrence and removal of antibiotic resistance genes in municipal wastewater and rural domestic sewage treatment systems in Eastern China. Environ Int 55(4):9–14. https://doi.org/10.1016/j.envint.2013.01.019
Cheng WX, Chen H, Su C, Yan SH (2013) Abundance and persistence of antibiotic resistance genes in livestock farms: a comprehensive investigation in Eastern China. Environ Int 61(4):1–7. https://doi.org/10.1016/j.envint.2013.08.023
Chopra I, Roberts M (2001) Tetracycline antibiotics: mode of action, applications, molecular biology, and epidemiology of bacterial resistance. Microbiol Mol Biol Rev 65(2):232–260. https://doi.org/10.1016/S0022-3093(98)00783-2
De Sotto RB, Medriano CA, Salcedo DE, Lee H, Cho Y, Kim S (2016) Effects of solids retention time on the fate of tetracycline resistance in SBRs for the treatment of carcass leachate. J Environ Manag 181:298–303. https://doi.org/10.1016/j.jenvman.2016.06.046
Ding HJ, Qiao M, Zhong JY, Zhu YG, Guo CJ, Zhang QQ, Yang P, Han L, Zhang WH, Wu YX, Liu JT, Zhang LT, Sun JH (2020) Characterization of antibiotic resistance genes and bacterial community in selected municipal and industrial sewage treatment plants beside Poyang Lake. Water Res 174:115603. https://doi.org/10.1016/j.watres.2020.115603
Duan ML, Gu J, Wang XJ, Li Y, Zhang RR, Hu T, Zhou BB (2019) Factors that affect the occurrence and distribution of antibiotic resistance genes in soils from livestock and poultry farms. Ecotoxicol Environ Saf 180:114–122. https://doi.org/10.1016/j.ecoenv.2019.05.005
Ebomah KE, Okoh AI (2020) An African perspective on the prevalence, fate and effects of carbapenem resistance genes in hospital effluents and wastewater treatment plant (WWTP) final effluents: a critical review. Heliyon 6(e03899). https://doi.org/10.1016/j.heliyon.2020.e03899
Etemadi S, Ebrahimzadeh Leylabadlo H, Ghotaslou R (2020) AmpC β-lactamase among Enterobacteriaceae: a new insight. Gene Rep 19:100673. https://doi.org/10.1016/j.genrep.2020.100673
Everage TJ, Boopathy R, Nathaniel R, Lafleur G, Doucet J (2014) A survey of antibiotic-resistant bacteria in a sewage treatment plant in thibodaux, Louisiana, USA. Int Biodeter Biodegrad 95(2):2–10. https://doi.org/10.1016/j.ibiod.2014.05.028
Gu JD (2019) Microbial ecotoxicology as an emerging research subject. Appl Environ Biotechnol. https://doi.org/10.26789/AEB.2019.01.001
Guo XP, Li J, Yang F, Yang J, Yin DQ (2014) Prevalence of sulfonamide and tetracycline resistance genes in drinking water treatment plants in the Yangtze River Delta, China. Sci Total Environ 493:626–631. https://doi.org/10.1016/j.scitotenv.2014.06.035
Harnisz M, Kiedrzynska E, Kiedrzynski M, Korzeniewska E, Czatzkowska M, Koniuszewska I, Jozwik A, Szklarek S, Niestepski S, Zalewski M (2020) The impact of WWTP size and sampling season on the prevalence of antibiotic resistance genes in wastewater and the river system. Sci Total Environ 741:140466. https://doi.org/10.1016/j.scitotenv.2020.140466
He P, Wu Y, Huang WZ, Wu XW, Lv JY, Liu PD, Bu L, Bai ZJ, Chen SY, Feng WR, Yang ZC (2020) Characteristics of and variation in airborne ARGs among urban hospitals and adjacent urban and suburban communities: a metagenomic approach. Environ Int 139:105625. https://doi.org/10.1016/j.envint.2020.105625
He XL, Xu YB, Chen JL, Ling JY, Li YF, Zhou X, Zheng L, Xie GY (2017) Evolution of corresponding resistance genes in the water of fish tanks with multiple stresses of antibiotics and heavy metals. Water Res 124:39–48. https://doi.org/10.1016/j.watres.2017.07.048
Huang L, Xu YB, Xu JX, Ling JY, Chen JL, Zhou JL, Zheng L, Du QP (2017) Antibiotic resistance genes (ARGs) in duck and fish production ponds with integrated or non-integrated mode. Chemosphere 168:1107–1114. https://doi.org/10.1016/j.chemosphere.2016.10.096
Huang MH, Zhang W, Liu C, Hu HY (2015) Fate of trace tetracycline with resistant bacteria and resistance genes in an improved AAO wastewater treatment plant. Process Saf Environ Protection 93:68–74. https://doi.org/10.1016/j.psep.2014.04.004
Huang W, Gong BZ, Wang YM, Lin ZY, He L, Zhou J, He Q (2020) Metagenomic analysis reveals enhanced nutrients removal from low C/N municipal wastewater in a pilot-scale modified AAO system coupling electrolysis. Water Res 173:115530. https://doi.org/10.1016/j.watres.2020.115530
Hu YR, Zhang TY, Jiang L, Luo Y, Yao SJ, Zhang D, Lin KF, Cui CZ (2019) Occurrence and reduction of antibiotic resistance genes in conventional and advanced drinking water treatment processes. Sci Total Environ 669:777–784. https://doi.org/10.1016/j.scitotenv.2019.03.143
Ibrahimagić A, Bedenić B, Kamberović F, Uzunović S (2015) High prevalence of CTX-M-5 and first report of CTX-M-3, CTX-M-22, CTX-M-28 and plasmid-mediated AmpC beta-lactamase producing Enterobacteriaceae, causing urinary tract infections in Bosnia and Herzegovina in hospital and community settings. J Infect Chemother 21(5):363–369. https://doi.org/10.1016/j.jiac.2015.01.003
Jacoby GA (2009) AmpC beta-lactamases. Clin Microbiol Rev 22(1):161. https://doi.org/10.1128/CMR.00036-08
Jang HM, Lee J, Shin SG, Shin J, Kim YM (2020) Comparing the fate of antibiotic resistance genes in two full-scale thermophilic anaerobic digestion plants treating food wastewater. Bioresource Technol 312:123577. https://doi.org/10.1016/j.biortech.2020.123577
Jaranowska P, Cydzik-Kwiatkowska A, Zielińska M (2013) Configuration of biological wastewater treatment line and influent composition as the main factors driving bacterial community structure of activated sludge. World J Microbiol Biotechnol 29(7):1145–1153. https://doi.org/10.1007/s11274-013-1273-9
Kim C, Ryu HD, Chung EG, Kim Y, Lee JK (2018) A review of analytical procedures for the simultaneous determination of medically important veterinary antibiotics in environmental water: sample preparation, liquid chromatography, and mass spectrometry. J Environ Manag 217:629–645. https://doi.org/10.1016/j.jenvman.2018.04.006
Korzeniewska E, Harnisz M (2013) Extended-spectrum beta-lactamase (ESBL)-positive Enterobacteriaceae in municipal sewage and their emission to the environment. J Environ Manag 128(20):904–911. https://doi.org/10.1016/j.jenvman.2013.06.051
Kumar M, Ram B, Sewwandi H, Sulfikar, Honda R, Chaminda T (2020) Treatment enhances the prevalence of antibiotic-resistant bacteria and antibiotic resistance genes in the wastewater of Sri Lanka, and India. Environ Res 183:109179. https://doi.org/10.1016/j.envres.2020.109179
Laht M, Karkman A, Voolaid V, Ritz C, Tenson T, Virta M, Kisand V (2014) Abundances of tetracycline, sulphonamide and beta-lactam antibiotic resistance genes in conventional wastewater treatment plants (WWTPs) with different waste load. Plos One 9(8):e103705. https://doi.org/10.1371/journal.pone.0103705
Lee J, Jeon JH, Shin J, Jang HM, Kim S, Song MS, Kim YM (2017) Quantitative and qualitative changes in antibiotic resistance genes after passing through treatment processes in municipal wastewater treatment plants. Sci Total Environ 605-606:906–914. https://doi.org/10.1016/j.scitotenv.2017.06.250
Li N, Sheng GP, Lu YZ, Zeng RJ, Yu HQ (2017) Removal of antibiotic resistance genes from wastewater treatment plant effluent by coagulation. Water Res 111:204–212. https://doi.org/10.1016/j.watres.2017.01.010
Li S, Zhang RJ, Hu JR, Shi WZ, Kuang YZ, Guo XY, Sun WL (2019) Occurrence and removal of antibiotics and antibiotic resistance genes in natural and constructed riverine wetlands in Beijing, China. Sci Total Environ 664:546–553. https://doi.org/10.1016/j.scitotenv.2019.02.043
Li Z, Li M, Zhang ZY, Li P, Zang YG, Liu Z (2020) Antibiotics in aquatic environments of China: a review and meta-analysis. Ecotoxicol Environ Saf 199:110668. https://doi.org/10.1016/j.ecoenv.2020.110668w
Li ZH, Yuan L, Gao SX, Wang L, Sheng GP (2019) Mitigated membrane fouling and enhanced removal of extracellular antibiotic resistance genes from wastewater effluent via an integrated pre-coagulation and microfiltration process. Water Res 159:145–152. https://doi.org/10.1016/j.watres.2019.05.005
Liu H, Sun HF, Zhang M, Liu Y (2019) Dynamics of microbial community and tetracycline resistance genes in biological nutrient removal process. J Environ Manag 238:84–91. https://doi.org/10.1016/j.jenvman.2019.02.123
Liu TK, Lun JS, Zheng P, Feng JR, Meng SS, Peng T, Hu Z (2020) Diversity and distribution of antibiotics and antibiotic resistance genes in seven national mangrove nature reserves, South China. Int Biodeter Biodegrad 153:105000. https://doi.org/10.1016/j.ibiod.2020.105000
Liu Y, Li JX, Guo WS, Ngo HH, Hu JJ, Gao MT (2018) Use of magnetic powder to effectively improve the performance of sequencing batch reactors (SBRs) in municipal wastewater treatment. Bioresour Technol 248:135–139. https://doi.org/10.1016/j.biortech.2017.06.069
Liu ZZ, Zhang JQ, Rao ST, Sun L, Zhang J, Liu R, Zheng GS, Ma XB, Hou SY, Zhuang XG, Song XY, Li QG (2015) Heptaplex PCR melting curve analysis for rapid detection of plasmid-mediated AmpC β-lactamase genes. J Microbiol Methods 110:1–6. https://doi.org/10.1016/j.mimet.2014.12.019
Mahrouki S, Chihi H, Bourouis A, Ayari K, Ferjani M, Moussa MB, Belhadj O (2015) Nosocomial dissemination of plasmids carrying blaTEM-24, blaDHA-1, aac (6’)-Ib-cr, and qnrA6 in Providencia, spp. strains isolated from a Tunisian hospital. Diagn Microbiol Infect Dis 81(1):50–52. https://doi.org/10.1016/j.diagmicrobio.2014.09.021
Maravić A, Skočibušić M, Šamanić I, Fredotović Ž, Cvjetan S, Jutronić M, Puizina J (2013) Aeromonas spp. simultaneously harbouring blaCTX-M-15, blaSHV-12, blaPER-1 and blaFOX-2, in wild-growing mediterranean mussel (Mytilus galloprovincialis) from Adriatic Sea, Croatia. Int J Food Microbiol 166(2):301–308. https://doi.org/10.1016/j.ijfoodmicro.2013.07.010
Mckinney CW, Loftin KA, Meyer MT, Davis JG, Pruden A (2010) Tet and sul antibiotic resistance genes in livestock lagoons of various operation type, configuration, and antibiotic occurrence. Environ Sci Technol 44(16):6102–6109. https://doi.org/10.1021/es9038165
Mckinney CW, Pruden A (2012) Ultraviolet disinfection of antibiotic resistant bacteria and their antibiotic resistance genes in water and wastewater. Environ Sci Technol 46(24):13393–13400. https://doi.org/10.1021/es303652q
McInnes RS, McCallum GE, Lamberte LE, van Schaik W (2020) Horizontal transfer of antibiotic resistance genes in the human gut microbiome. Curr Opin Microbiol 53:35–43. https://doi.org/10.1016/j.mib.2020.02.002
Medema G, Heijnen L, Elsinga G, Italiaander R, Brouwer A (2020) Presence of SARS-Coronavirus-2 in Sewage. medRxiv. https://doi.org/10.1101/2020.03.29.20045880
Mohamudha PR, Harish BN, Parija SC (2012) Molecular description of plasmid-mediated AmpC β-lactamases among nosocomial isolates of Escherichia coli & Klebsiella pneumoniae from six different hospitals in India. Indian J Med Res 135(1):114–119. https://doi.org/10.4103/0971-5916.93433
Nõlvak H, Truu M, Tiirik K, Oopkaup K, Sildvee T, Kaasik A, Mander Ü, Truu J (2013) Dynamics of antibiotic resistance genes and their relationships with system treatment efficiency in a horizontal subsurface flow constructed wetland. Sci Total Environ 462(7):636–644. https://doi.org/10.1016/j.scitotenv.2013.05.052
Organization WH (2014) Antimicrobial resistance: global report on surveillance. Australasian Med J 7(4):695–704. https://doi.org/10.2307/2342307
Pallares-Vega R, Blaak H, van der Plaats R, de Roda Husman AM, Hernandez Leal L, van Loosdrecht MCM, Weissbrodt DG, Schmitt H (2019) Determinants of presence and removal of antibiotic resistance genes during WWTP treatment: a cross-sectional study. Water Res 161:319–328. https://doi.org/10.1016/j.watres.2019.05.100
Pazda M, Kumirska J, Stepnowski P, Mulkiewicz E (2019) Antibiotic resistance genes identified in wastewater treatment plant systems-A review. Sci Total Environ 697:134023. https://doi.org/10.1016/j.scitotenv.2019.134023
Qin KN, Wei LL, Li JJ, Lai B, Zhu FY, Yu H, Zhao QL, Wang K (2020) A review of ARGs in WWTPs: Sources, stressors and elimination. Chin Chem Lett. https://doi.org/10.1016/j.cclet.2020.04.057
Riquelme Breazeal MV, Novak JT, Vikesland PJ, Pruden A (2013) Effect of wastewater colloids on membrane removal of antibiotic resistance genes. Water Res 47(1):130–140. https://doi.org/10.1016/j.watres.2012.09.044
Sharma VK, Johnson N, Cizmas L, McDonald TJ, Kim H (2016) A review of the influence of treatment strategies on antibiotic resistant bacteria and antibiotic resistance genes. Chemosphere 150:702–714. https://doi.org/10.1016/j.chemosphere.2015.12.084
Shi P, Jia SY, Zhang XX, Zhang T, Cheng SP, Li AM (2013) Metagenomic insights into chlorination effects on microbial antibiotic resistance in drinking water. Water Res 47(1):111–120. https://doi.org/10.1016/j.watres.2012.09.046
Souissi M, Laabidi R, Aissa P, Pringault O, Said OB (2018) Influence of Bizerte city wastewater treatment plant (WWTP) on abundance and antibioresistance of culturable heterotrophic and fecal indicator bacteria of Bizerte Lagoon (Tunisia). Ecotoxicol Environ Saf 148:201–210. https://doi.org/10.1016/j.ecoenv.2017.10.002
Storteboom HN, Kim SC, Doesken KC, Carlson KH, Davis JG, Pruden A (2007) Response of antibiotics and resistance genes to high-intensity and low-intensity manure management. J Environ Qual 36(6):1695–1703. https://doi.org/10.2134/jeq2007.0006
Su HC, Ying GG, He LY, Liu YS, Zhang RQ, Tao R (2014) Antibiotic resistance, plasmid-mediated quinolone resistance (PMQR) genes and AmpC gene in two typical municipal wastewater treatment plants. Environ Sci Processes Impacts 16(2):324–332. https://doi.org/10.1039/c3em00555k
Su YL, Wang JX, Xia HP, Xie B, Li X (2019) Anaerobic/aerobic conditions determine antibiotic resistance genes removal patterns from leachate by affecting bacteria taxa-genes cooccurrence modules. Chemosphere 223:28–38. https://doi.org/10.1016/j.chemosphere.2019.02.007
Tao CW, Hsu BM, Ji WT, Hsu TK, Kao PM, Hsu CP, Shen SM, Shen TY, Wan TJ, Huang YL (2014) Evaluation of five antibiotic resistance genes in wastewater treatment systems of swine farms by real-time PCR. Sci Total Environ 496:116–121. https://doi.org/10.1016/j.scitotenv.2014.07.024
Venieri D, Gounaki I, Bikouvaraki M, Binas V, Zachopoulos A, Kiriakidis G, Mantzavinos D (2017) Solar photocatalysis as disinfection technique: inactivation of Klebsiella pneumoniae in sewage and investigation of changes in antibiotic resistance profile. J Environ Manag 195:140–147. https://doi.org/10.1016/j.jenvman.2016.06.009
Wang CY, Dang HY, Ding YS (2008) Incidence of diverse integrons and β-lactamase genes in environmental Enterobacteriaceae isolates from Jiaozhou bay, China. World J Microbiol Biotechnol 24(12):2889–2896. https://doi.org/10.1007/s11274-008-9827-y
Wang HC, Wang J, Li SM, Ding GY, Wang K, Zhuang T, Huang X, Wang XY (2020) Synergistic effect of UV/chlorine in bacterial inactivation, resistance gene removal, and gene conjugative transfer blocking. Water Res 185:116290. https://doi.org/10.1016/j.watres.2020.116290
Wang JL, Mao DQ, Mu QH, Luo Y (2015) Fate and proliferation of typical antibiotic resistance genes in five full-scale pharmaceutical wastewater treatment plants. Sci Total Environ 526:366–73. https://doi.org/10.1016/j.scitotenv.2015.05.046
Widyasari-Mehta A, Hartung S, Kreuzig R (2016) From the application of antibiotics to antibiotic residues in liquid manures and digestates: a screening study in one European center of conventional pig husbandry. J Environ Manag 177:129–137. https://doi.org/10.1016/j.jenvman.2016.04.012
Wu DL, Zhang M, He LX, Zou HY, Liu YS, Li BB, Yang YY, Liu CX, He LY, Ying GG (2020) Contamination profile of antibiotic resistance genes in ground water in comparison with surface water. Sci Total Environ 715:136975. https://doi.org/10.1016/j.scitotenv.2020.136975
Xu R, Zhang YR, Xiong WP, Sun WM, Fan Q, Yang ZH (2020) Metagenomic approach reveals the fate of antibiotic resistance genes in a temperature-raising anaerobic digester treating municipal sewage sludge. J Clean Prod 277:123504. https://doi.org/10.1016/j.jclepro.2020.123504
Xu YB, Hou MY, Li YF, Huang L, Ruan JJ, Zheng L, Qiao QX, Du QP (2017) Distribution of tetracycline resistance genes and AmpC β-lactamase genes in representative non-urban sewage plants and correlations with treatment processes and heavy metals. Chemosphere 170:274–281. https://doi.org/10.1016/j.chemosphere.2016.12.027
Yan J, Xie JH, Wang SJ, Zhang HG, Wu JP, Hong YG (2019) Evaluation of nitrogen removal processes and microbial communities in eight full-scale municipal wastewater treatment plants. Appl Environ Biotechnol 4(1):49–59. https://doi.org/10.26789/AEB.2019.01.007
Yang YY, Xu C, Cao XH, Lin H, Wang J (2017) Antibiotic resistance genes in surface water of eutrophic urban lakes are related to heavy metals, antibiotics, lake morphology and anthropic impact. Ecotoxicology 26:831–840. https://doi.org/10.1007/s10646-017-1814-3
Ye Y, Xu XH, Li JB (2010) Emergence of CTX-M-3, TEM-1 and a new plasmid-Mediated MOX-4 AmpC in a multiresistant Aeromonas caviae isolate from a patient with pneumonia. J Med Microbiol 59(7):843–847. https://doi.org/10.1099/jmm.0.016337-0
Yuan QB, Guo MT, Wei WJ, Yang J (2016) Reductions of bacterial antibiotic resistance through five biological treatment processes treated municipal wastewater. Environ Sci Pollut Res 23(19):1–9. https://doi.org/10.1007/s11356-016-7048-8
Yuan QB, Zhai YF, Mao BY, Hu N (2018) Antibiotic resistance genes and intI1 prevalence in a swine wastewater treatment plant and correlation with metal resistance, bacterial community and wastewater parameters. Ecotoxicol Environ Saf 161:251–259. https://doi.org/10.1016/j.ecoenv.2018.05.049
Zhang HH, He HY, Chen SN, Huang TL, Lu KY, Zhang ZH, Wang R, Zhang XY, Li HL (2019) Abundance of antibiotic resistance genes and their association with bacterial communities in activated sludge of wastewater treatment plants: geographical distribution and network analysis. J Environ Sci 82:24–38. https://doi.org/10.1016/j.jes.2019.02.023
Zhang JY, Yang M, Zhong H, Liu MM, Sui QW, Zheng LB, Tong J, Wei YS (2018) Deciphering the factors influencing the discrepant fate of antibiotic resistance genes in sludge and water phases during municipal wastewater treatment. Bioresour Technol 265:310–319. https://doi.org/10.1016/j.biortech.2018.06.021
Zhang K, Niu ZG, Lv ZW, Zhang Y (2017) Occurrence and distribution of antibiotic resistance genes in water supply reservoirs in Jingjinji area, China. Ecotoxicology 26:1284–1292. https://doi.org/10.1007/s10646-017-1853-9
Zhang XX, Zhang T (2011) Occurrence, abundance, and diversity of tetracycline resistance genes in 15 sewage treatment plants across China and other global locations. Environ Sci Technol 45(7):2598–2604. https://doi.org/10.1021/es103672x
Zhang YH, Ma QP, Su BM, Chen R, Lin J, Lin ZF, Wang DL, Yu Y (2018) A study on the role that quorum sensing play in antibiotic-resistant plasmid conjugative transfer in Escherichia coli. Ecotoxicology 27(2):209–216. https://doi.org/10.1007/s10646-017-1886-0
Zhang YY, Zhuang Y, Geng JJ, Ren HQ, Zhang Y, Ding LL, Xu K (2015) Inactivation of antibiotic resistance genes in municipal wastewater effluent by Chlorination and sequential UV/Chlorination disinfection. Sci Total Environ 512-513(9):125–132. https://doi.org/10.1016/j.scitotenv.2015.01.028
Zhou CS, Wu JW, Dong LL, Liu BF, Xing DF, Yang SS, Wu XK, Wang Q, Fan JN, Feng LP, Cao GL (2020) Removal of antibiotic resistant bacteria and antibiotic resistance genes in wastewater effluent by UV-activated persulfate. J Hazard Mater 388:122070. https://doi.org/10.1016/j.jhazmat.2020.122070
Zhou M, Xu YB, Ouyang PQ, Ling JY, Cai QJ, Huang L, Zhou X (2020) Evolution and distribution of resistance genes and bacterial community in water and biofilm of a simulated fish-duck integrated pond with stress. Chemosphere 245:125549. https://doi.org/10.1016/j.chemosphere.2019.125549
Zhou S, Zhu YJ, Yan Y, Wang WG, Wang YY (2019) Deciphering extracellular antibiotic resistance genes (eARGs) in activated sludge by metagenome. Water Res 161:610–620. https://doi.org/10.1016/j.watres.2019.06.048
Funding
This work was financially supported by Nature Science Foundation of China (No. 41671481; 41977340), Key-Area Research and Development Program of Guangdong Province (2019B110205004) and National Key R & D Program of China (No.2016YFC0400706).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Lin, X., Ruan, J., Huang, L. et al. Comparison of the elimination effectiveness of tetracycline and AmpC β-lactamase resistance genes in a municipal wastewater treatment plant using four parallel processes. Ecotoxicology 30, 1586–1597 (2021). https://doi.org/10.1007/s10646-020-02306-0
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10646-020-02306-0