Abstract
Background
Pseudogenes are defined as key regulators in cancer initiation and progression. But their biological function and clinical significance in hepatocellular carcinoma (HCC) remain to be elucidated. In the current study, we identified a novel pseudogene, Annexin A2 pseudogene 1 (ANXA2P1), in HCC and explored its underlining molecular mechanism.
Methods and Results
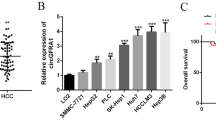
We analyzed the expression pattern of ANXA2P1 in a TCGA dataset and an HCC sample cohort and evaluated its clinical significance. The biological effects on HCC cells proliferation, migration, invasion and epithelial-mesenchymal transition (EMT) process were assessed by Cell Counting Kit-8 assay, Transwell assay and Western blot, respectively. The ANXA2P1/miR-376a-3p/ANXA2 axis was determined by bioinformatics analysis and dual-luciferase reporter assays. ANXA2P1 exerted as an oncogene that was significantly overexpressed in HCC tissues and was associated with disease progression and unfavorable prognosis of HCC patients. ANXA2P1 knockdown suppressed cell growth, cell migration and invasion and reversed EMT phenotype in HCC. Mechanistically, ANXA2P1 acts as a competing endogenous RNA for miR-376a-3p, thereby leading to the upregulation of its cognate gene ANXA2.
Conclusions
ANXA2P1/miR-376a-3p/ANXA2 axis plays an important role in the progression of HCC. Our findings may provide valuable therapeutic target for treating HCC.







Similar content being viewed by others
Availability of data and materials
All data that support the findings of this study are available from the corresponding authors upon reasonable request.
Abbreviations
- HCC:
-
Hepatocellular carcinoma
- ANXA2P1:
-
Annexin A2 pseudogene 1
- EMT:
-
Epithelial-mesenchymal transition
- RPMI:
-
Roswell Park Memorial Institute
- DMEM:
-
Dulbecco’s Modified Eagle Medium
- CCK-8:
-
Cell Counting Kit 8
- lncRNAs:
-
Long non-coding RNAs
- PTENP1:
-
PTEN tumor suppressor
- qRT-PCR:
-
Quantitative real-time polymerase chain reaction
- EdU:
-
5-Ethynyl-2′-deoxyuridine
- TCGA:
-
The Cancer Genome Atlas
- BP:
-
Biological process
- CC:
-
Cellular component
- MF:
-
Molecular function
References
Forner A, Reig M, Bruix J. Hepatocellular carcinoma. Lancet. 2018;391:1301–1314. https://doi.org/10.1016/S0140-6736(18)30010-2.
Maluccio M, Covey A. Recent progress in understanding, diagnosing, and treating hepatocellular carcinoma. CA Cancer J Clin. 2012;62:394–399. https://doi.org/10.3322/caac.21161.
Chen W, Sun K, Zheng R, et al. Cancer incidence and mortality in China, 2014. Chin J Cancer Res. 2018;30:1–12. https://doi.org/10.21147/j.issn.1000-9604.2018.01.01.
Esteller M. Non-coding RNAs in human disease. Nat Rev Genet. 2011;12:861–874. https://doi.org/10.1038/nrg3074.
Cech TR, Steitz JA. The noncoding RNA revolution-trashing old rules to forge new ones. Cell. 2014;157:77–94. https://doi.org/10.1016/j.cell.2014.03.008.
Milligan MJ, Lipovich L. Pseudogene-derived lncRNAs: emerging regulators of gene expression. Front Genet. 2014;5:476. https://doi.org/10.3389/fgene.2014.00476.
Poliseno L, Salmena L, Zhang J, Carver B, Haveman WJ, Pandolfi PP. A coding-independent function of gene and pseudogene mRNAs regulates tumour biology. Nature. 2010;465:1033–1038. https://doi.org/10.1038/nature09144.
Johnsson P, Ackley A, Vidarsdottir L, et al. A pseudogene long-noncoding-RNA network regulates PTEN transcription and translation in human cells. Nat Struct Mol Biol. 2013;20:440–446. https://doi.org/10.1038/nsmb.2516.
Qian YY, Li K, Liu QY, Liu ZS. Long non-coding RNA PTENP1 interacts with miR-193a-3p to suppress cell migration and invasion through the PTEN pathway in hepatocellular carcinoma. Oncotarget. 2017;8:107859–107869. https://doi.org/10.18632/oncotarget.22305.
Shi H, Xiao L, Duan W, et al. ANXA2 enhances the progression of hepatocellular carcinoma via remodeling the cell motility associated structures. Micron. 2016;85:26–33. https://doi.org/10.1016/j.micron.2016.03.008.
Chandrashekar DS, Bashel B, Balasubramanya SAH, et al. UALCAN: a portal for facilitating tumor subgroup gene expression and survival analyses. Neoplasia. 2017;19:649–658. https://doi.org/10.1016/j.neo.2017.05.002.
Cerami E, Gao J, Dogrusoz U, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–404. https://doi.org/10.1158/2159-8290.CD-12-0095.
Gao J, Aksoy BA, Dogrusoz U, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6:l1. https://doi.org/10.1126/scisignal.2004088.
Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Prot. 2009;4:44.
Li JH, Liu S, Zhou H, Qu LH, Yang JH. starBase v2.0: decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA interaction networks from large-scale CLIP-Seq data. Nucleic Acids Res. 2014;42:92–97. https://doi.org/10.1093/nar/gkt1248.
Basu S, Cheriyamundath S, Ben-Ze’ev A. Cell–cell adhesion: linking Wnt/beta-catenin signaling with partial EMT and stemness traits in tumorigenesis. F1000Res. 2018;. https://doi.org/10.12688/f1000research.15782.1.
An Y, Furber KL, Ji S. Pseudogenes regulate parental gene expression via ceRNA network. J Cell Mol Med. 2017;21:185–192. https://doi.org/10.1111/jcmm.12952.
Zhang Z, Gerstein M. Large-scale analysis of pseudogenes in the human genome. Curr Opin Genet Dev. 2004;14:328–335. https://doi.org/10.1016/j.gde.2004.06.003.
Xiao-Jie L, Ai-Mei G, Li-Juan J, Jiang X. Pseudogene in cancer: real functions and promising signature. J Med Genet. 2015;52:17–24. https://doi.org/10.1136/jmedgenet-2014-102785.
Yang W, Du WW, Li X, Yee AJ, Yang BB. Foxo3 activity promoted by non-coding effects of circular RNA and Foxo3 pseudogene in the inhibition of tumor growth and angiogenesis. Oncogene. 2016;35:3919–3931. https://doi.org/10.1038/onc.2015.460.
Esposito F, De Martino M, Forzati F, Fusco A. HMGA1-pseudogene overexpression contributes to cancer progression. Cell Cycle. 2014;13:3636–3639. https://doi.org/10.4161/15384101.2014.974440.
Hayashi H, Arao T, Togashi Y, et al. The OCT4 pseudogene POU5F1B is amplified and promotes an aggressive phenotype in gastric cancer. Oncogene. 2015;34:199–208. https://doi.org/10.1038/onc.2013.547.
Yu G, Yao W, Gumireddy K, et al. Pseudogene PTENP1 functions as a competing endogenous RNA to suppress clear-cell renal cell carcinoma progression. Mol Cancer Ther. 2014;13:3086–3097. https://doi.org/10.1158/1535-7163.MCT-14-0245.
Zhu Q, Liu J, Tang J, Guo DL, Li Y, Duan R. Overexpression of long non-coding RNAs DUXAP9 and DUXAP10 is associated with prognosis in patients with hepatocellular carcinoma after hepatectomy. Int J Clin Exp Pathol. 2018;11:1407–1414.
Kong Y, Zhang L, Huang Y, et al. Pseudogene PDIA3P1 promotes cell proliferation, migration and invasion, and suppresses apoptosis in hepatocellular carcinoma by regulating the p53 pathway. Cancer Lett. 2017;407:76–83. https://doi.org/10.1016/j.canlet.2017.07.031.
Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. https://doi.org/10.1016/j.cell.2011.02.013.
He C, Zhou Z, Jiang H, et al. Epithelial-mesenchymal transition is superior to vessels-encapsulate tumor cluster in promoting metastasis of hepatocellular carcinoma: a morphological evidence. J Cancer. 2017;8:39–47. https://doi.org/10.7150/jca.16736.
Brabletz T, Kalluri R, Nieto MA, Weinberg RA. EMT in cancer. Nat Rev Cancer. 2018;18:128–134. https://doi.org/10.1038/nrc.2017.118.
Tam WL, Weinberg RA. The epigenetics of epithelial-mesenchymal plasticity in cancer. Nat Med. 2013;19:1438–1449. https://doi.org/10.1038/nm.3336.
Muro EM, Mah N, Andrade-Navarro MA. Functional evidence of post-transcriptional regulation by pseudogenes. Biochimie. 2011;93:1916–1921. https://doi.org/10.1016/j.biochi.2011.07.024.
Zhang X, Liu S, Guo C, Zong J, Sun MZ. The association of annexin A2 and cancers. Clin Transl Oncol. 2012;14:634–640. https://doi.org/10.1007/s12094-012-0855-6.
Acknowledgment
We would like to give our sincere appreciation to the reviewers for their helpful comments on this article.
Funding
This study was funded by the Scientific Research Starting Program of Shunde Hospital, Southern Medical University (The First People’s Hospital of Shunde, Foshan) (SRSP2019016) and Medical Scientific Research Foundation of Guangdong Province of China (2017113015361).
Author information
Authors and Affiliations
Contributions
Weidong Wang designed and supervised the study. Huohui Ou and Qingbo Liu performed the experiments and acquired result data. Jie Lin and Wei He collected the patient samples and information. Weijie Zhang and Jing Ma helped to review the statistical analysis. Huohui Ou drafted the manuscript. Weidong Wang and Qingbo Liu critically revised the manuscript. All authors have read and approved the final version of the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors report no conflicts of interest in this work.
Consent for publication
Not applicable.
Ethics approval and consent to participate
This animal research was approved by the Ethical Committee of Shunde Hospital, Southern Medical University (The First People’s Hospital of Shunde).
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic Supplementary Material
Below is the link to the electronic supplementary material.

Figure S1.
Expression of ANXA2P1 in TCGA samples cohort. (JPEG 68 kb)

Figure S2.
The positive correlation between ANXA2P1 and ANXA2 in TCGA samples cohort. (JPEG 94 kb)
Rights and permissions
About this article
Cite this article
Ou, H., Liu, Q., Lin, J. et al. Pseudogene Annexin A2 Pseudogene 1 Contributes to Hepatocellular Carcinoma Progression by Modulating Its Parental Gene ANXA2 via miRNA-376a-3p. Dig Dis Sci 66, 3903–3915 (2021). https://doi.org/10.1007/s10620-020-06734-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10620-020-06734-0