Abstract
The k-means algorithm is widely used for clustering, compressing, and summarizing vector data. We present a fast and memory-efficient GPU-based algorithm for exact k-means, Asynchronous Selective Batched K-means (ASB K-means). Unlike most GPU-based k-means algorithms that require loading the whole dataset onto the GPU for clustering, the amount of GPU memory required to run our algorithm can be chosen to be much smaller than the size of the whole dataset. Thus, our algorithm can cluster datasets whose size exceeds the available GPU memory. The algorithm works in a batched fashion and applies the triangle inequality in each k-means iteration to omit a data point if its membership assignment, i.e., the cluster it belongs to, remains unchanged, thus significantly reducing the number of data points that need to be transferred between the CPU’s RAM and the GPU’s global memory and enabling the algorithm to very efficiently process large datasets. Our algorithm can be substantially faster than a GPU-based implementation of standard k-means even in situations when application of the standard algorithm is feasible because the whole dataset fits into GPU memory. Experiments show that ASB K-means can run up to 15x times faster than a standard GPU-based implementation of k-means, and it also outperforms the GPU-based k-means implementation in NVIDIA’s open-source RAPIDS machine learning library on all the datasets used in our experiments.
Similar content being viewed by others
1 Introduction
Advances in science and technologies cause data to accumulate at an explosive speed (Fahad et al. 2014; Sajana et al. 2016). For example, an enormous volume of data has been collected by electronic devices and sensors in IoT networks, and online social networks such as Twitter, Tumblr, and Weibo generate a huge amount of data every day. These datasets often contains useful information—for example, knowledge extracted from social media networks can be used to study public opinions, analyse consumer trends, and provide intelligent advertisement and content catering to the user’s specific interest—and how to efficiently extract it has attracted much interest in the realm of data mining.
In this paper, we consider clustering of large datasets using the classic k-means algorithm. Clustering is the process of partitioning data elements in a dataset so that similar ones are collected in the same group (or cluster). It is applicable in many domains and to many types of data (Berkhin 2006). Compared with the original data, clusters can be digested by humans far more effectively and efficiently. As an unsupervised learning task, clustering is applicable in many fields, including image and video processing, social science, energy studies, bioinformatics, biochemistry, and marketing (Xu and Wunsch 2005; Mohebi et al. 2016). Therefore, a lot of research work has been done on new clustering algorithms in the past fifty years (Jain 2010). However, in spite of its age, the classic k-means algorithm is still used very widely, perhaps primarily due to its conceptual simplicity.
Batch clustering algorithms such as the standard k-means algorithm generally examine every data element of a dataset several times to find locally or globally optimal solutions, rendering them computationally more expensive than one-pass algorithms and time-consuming to run on very large datasets. Parallel computing has proven to be a practical solution for handling big data (Fang et al. 2008; Jian et al. 2013; Langdon 2013; Shirkhorshidi et al. 2014): many complex data mining tasks can be dramatically sped up by dividing the task into many sub-tasks and executing them concurrently. Traditionally, supercomputers are used for high-performance parallel computing and have been applied to solve complex scientific problems (Upadhyaya 2013). However, traditional supercomputers are expensive to acquire and run, greatly limiting their applicability. Graphics processing units (GPUs) offer an alternative. They were designed originally for computer graphics, where each pixel of the screen is processed by a separate processing element (thread), requiring massive parallelism. This, coupled with increasing programmability, makes them an attractive platform for general-purpose high-performance computing (Owens et al. 2008). GPUs have been utilised in various domains and applications such as scientific and geometric computations, matrix operations, FFT computation, embedded system design, bioinformatics, database operations, and data mining (Owens et al. 2005; Che et al. 2008; Brodtkorb et al. 2013; Mittal and Vetter 2015). In particular, they have been used to significantly accelerate algorithms for clustering problems (Li et al. 2013)—for example, by applying the general-purpose CUDA programming model on NVIDIA GPU architectures (Wu et al. 2009). However, existing GPU-based implementations of the popular k-means clustering algorithm are bound by the limited amount of GPU global memory and do not make use of geometric constraints to speed up the clustering process, which we address in this paper.
Our primary contributions can be summarized as follows:
-
We present the first GPU-based k-means algorithm that can handle datasets of any size and very large numbers of cluster centers.
-
Our method is the first GPU-based k-means algorithm that applies the triangle inequality to avoid redundant distance computations and reduce the amount of data that needs to be transferred between the CPU and GPU.
-
We demonstrates how CUDA’s asynchronous APIs can be used to effectively hide the overhead involved in transferring data between the CPU’s main memory and the GPU’s global memory.
The rest of the paper is structured in the following manner. We give a brief overview of prior work on k-means clustering and GPU-based k-means in Sect. 2. In Sect. 3, we present our batched GPU-based k-means algorithm accelerated by the triangle inequality and its implementation. Section 4 discusses the experimental results on different datasets using different k-means algorithms. The last section presents our conclusions.
2 Related work
First, we briefly review the background on general-purpose computation on GPUs before focusing on existing work comprising (a) CPU-based algorithms that accelerate k-means using the triangle inequality and (b) GPU-based implementations of k-means.
2.1 GPGPU
General-purpose computing with graphics processing units (GPGPU) is the utilisation of a GPU, which was originally designed for handling computation specifically for computer graphics, to carry out computation traditionally performed by the a CPU. GPGPU only became practical and popular after 2001 because of programmable vertex shaders and floating-point support introduced in modern graphics processors (Owens et al. 2005). Because GPUs devote more transistor space than CPUs to data processing rather than data caching and flow control, they are especially suitable for dealing with problems that can be expressed as data-parallel computations with high arithmetic intensity.
From the hardware perspective, and using the terminology employed by GPU manufacturer NVIDIA, a GPU consists of a grid of so-called streaming multiprocessors (SMPs), each of which consists of a set of simple scalar processors (SPs). These scalar processors, also called “cores”, operate in a single instruction, multiple data fashion (SIMD). Each SMP has its own set of registers, and each SMP has a shared memory block that all cores in the SMP, and only those cores, can access. Thus, when it comes to GPGPU, three main types of memory exist on GPUs: registers, shared memory, and global memory (a.k.a., device memory). Another two types of memory: constant and texture memory, are less used because they are beneficial for only very specific types of applications.
Registers are 32-bit memory blocks that are private to each processing core. Access to registers is extremely fast but their number is limited. Shared memory is on-chip memory with faster access than device memory. It is limited in size (e.g., 16, 32 or 48KB) and usually used as a local buffer for fast data retrieval and information exchange between threads running on the same SMP. Device memory is physically located on the graphics card but not inside the processing unit. It is also called global memory because it can be accessed from both the GPU and CPU (with the help of some specialised API functions). Device memory is much larger, say 4, 20, 40, or even 80GB, than shared memory and can be accessed by all threads on different SMPs. Because it is off-chip, the access latency is hundreds of clock cycles, which is much slower than accessing registers and shared memory. Therefore, appropriate programming techniques should be employed to help hide memory access latency. For example, tiled loading of data from device memory into shared memory and pipelining are two widely used techniques (NVIDIA 2021).
The CPU’s main memory located on the motherboard is normally large in size but can only be accessed by the CPU. Data has to be explicitly transferred from main memory to device memory before it can be processed by the GPU. Because the bandwidth of the PCI-Express bus used for data transfer is limited, this can easily become a bottleneck. The time incurred by transferring data between the main memory of the CPU and the device memory of the GPU is an extra cost relative to traditional data mining methods running on CPUs only. This is a hindrance that needs to be circumvented when mining large datasets on GPUs.
Threads are basic constructs of the GPGPU programming model. Because they are lightweight and incur very little creation and context switching overhead, they are often used to hide memory access latency. Threads are grouped into warps. All threads in a warp execute the same instructions on different data elements. Warps are further grouped into thread blocks. It is guaranteed that a thread block resides on one streaming multiprocessor. Threads in the same block can cooperate by sharing data through shared memory and their execution can be synchronised, but communication and synchronisation between thread blocks is limited. Thread blocks, in turn, are organised into a computing grid, which is processed jointly by the collection of streaming multiprocessors in the GPU.
2.2 K-means clustering
The k-means clustering algorithm is one of the most widely used clustering algorithms in both academic research and industrial practice. Given a user-specified number of clusters k, it partitions a set of n data points into k clusters according to a distance measure. The metric used to measure distance is usually defined as the sum of squared distances between each member in the cluster and the center (i.e., mean) of the cluster, called intra-cluster distance. In this scenario, which is the one we consider in this paper, the objective of k-means is to minimize the sum of all intra-cluster distances. This sum is also referred to as the sum of squared errors (SSE).
The standard implementation of k-means clustering is Lloyd’s algorithm (Lloyd 1982). It can be described as follows:
-
1.
The k centers are initialized according to some rules (e.g., selected at random from the data points). This is called the initialization step.
-
2.
Each data point is assigned to its closest centroid to form k clusters. This is referred to as the assignment step.
-
3.
All data points assigned to a given cluster are used to recalculate the centroid. This is called the recalculation step.
-
4.
The procedure (Steps 2 and 3) is repeated until a certain termination condition is reached. This is referred to as the termination step.
It is important to note that the algorithm will converge to a local minimum of the SSE but will not necessarily find a global minimum. Hence, it is common practice to restart the algorithm multiple times with different randomly chosen initializations.
Many termination conditions have been reported in the literature. The most commonly used ones include:
-
1.
A maximum number of iterations has been executed.
-
2.
The centroids remain unchanged, i.e., the centroid positions do not change at all or only change slightly in consecutive steps.
-
3.
The assignment of each data point does not change. This is equivalent to Condition 2 in most situations but enables a different implementation.
-
4.
The sum of squared errors (SSE) reaches the desired minimum value.
Researchers have developed several optimizations to improve on the basic k-means algorithm, generally by exploiting either algorithmic improvements or parallelization. Most of the algorithmic improvements are primarily concerned with optimizing the membership assignment step of the algorithm (that is, finding the closest center for each data point). With the advent of GPGPU, parallelization as an optimization technique has also been widely studied. We will briefly review these two optimization strategies in the next two sections. There is another direction of acceleration that considers approximation techniques, e.g., sub-sampling (Bejarano et al. 2011). In this paper, we consider exact techniques only.
2.3 Triangle inequality accelerated K-means clustering
Several variants of k-means speed up the clustering process by avoiding distance calculations between the clustered data points and cluster centers using the triangle inequality. All the optimizations covered in this section produce exactly the same results as Lloyd’s algorithm given the same input and initialization, so they are suitable as drop-in replacements.
Three observations can be made in the standard k-means algorithm (Elkan 2003):
-
1.
In later iterations, there is little movement of centers.
-
2.
Distance calculations use the most time.
-
3.
Geometrically, distance calculations are mostly redundant.
Lloyd’s algorithm spends a lot of processing time computing distances between each of the k cluster centers and the n data points to find the closest center for a data point. However, since points usually stay in the same cluster after a few iterations of the clustering algorithm, more often than not, much of this work is unnecessary and wasted. By applying the triangle inequality, computing the distance between a data point and any cluster center can be avoided if the center assignment is guaranteed to remain unchanged.
Using the triangle inequality, the following two propositions can be proved:
Proposition 1
If \( d(c, c') \ge 2d(x, c) \), then \( d(x, c') \ge d(x, c) \).
Proof
If
then, using the triangle inequality \( d(x, c) + d(x, c') \ge d(c, c') \), it follows that
that is,
This means that, in this situation, the data point x is closer to center c than \( c' \), and we do not need to measure the distance \( d(x, c') \). Assuming that the distances \( d(c, c') \) and d(x, c) have been cached, this is trivial to check. By applying the triangle inequality in this way, computation of the distance from data point x to center \( c' \) can be avoided.
Proposition 2
The distance from a data point x to a moved center c is bounded by the movement of the center.
Proof
Assuming the center c is moved to a new position \( c' \) after an iteration, given the distance of a data point x to c, i.e., d(x, c) , and the distance between the center c and the updated center \( c' \), i.e., \( d(c, c') \), we know from the triangle inequality that:
This means the distance from the data point x to the updated center \( c' \) must be inside the range bounded by \( d(x, c) - d(c, c') \), i.e., the lower bound, and \( d(x, c) + d(c, c') \), i.e., the upper bound. Again, assuming the distances d(x, c) and \( d(c, c') \) have been cached, it is trivial to calculate these bounds.
By applying Proposition 1 directly in the innermost loop of k-means, Phillips (2002) presents two accelerated algorithms named compare-means and sort-means. The algorithms compute and cache the center-point and center-center distances. If twice the distance of a data point x to its assigned center c(x) is smaller than the distance of the center c to another center \( c' \), then the data point x will not be assigned to the center \( c' \) in the next iteration. In this way, some center-point distance recomputations can be avoided.
Elkan (2003) presents an algorithm that uses both Propositions 1 and 2 to avoid redundant distance calculations. He introduces the concept of upper and lower bounds for distances between data points and centers, and demonstrates an effective way to speed up the standard k-means algorithm by maintaining 1 upper bound and k lower bounds for each data point. The upper bound for data point x, u(x) , is the greatest possible value of the distance from the data point x to its assigned center c(x) , while the lower bounds, l(x, j) , correspond to the least possible distances between the data point x and each center j. The pseudo-code in Algorithm 1 shows the workflow of Elkan’s algorithm.
By applying Proposition 1, Elkan proved that, given a data point x, its assigned center c, and another center \(c'\), if \( u(x) \le d(c, c') / 2 \), then x will stay closer to c than \(c'\) in the next iteration. Hence, the distance computation between data point x and center \( c' \) is unnecessary and can be avoided. If the upper bound u(x) is smaller than all the half-distances from center c to other centers, i.e., \( u(x) \le \min _{c' \in C;c' \ne c}(d(c, c') / 2) \) where C is the set of centers, then there is no need to calculate the distance of x to any center because its center assignment will not change.
By applying Proposition 2, Elkan also pointed out that, for a center, the calculation of the point-center distance can be avoided if the data point’s upper bound is less than or equal to the lower bound for that center. This can be proved as follows. Given two centers c and \( c' \), where c is the cluster center that point x is assigned to, if \( u(x) \le l(x, c') \), then \( d(x, c) \le u(x) \le l(x, c') \le d(x, c') \) (according to the definition of the upper and lower bounds), that is, \( d(x, c) \le d(x, c') \). This means that x stays closer to c than \( c' \), and there is no need to calculate the distance between x and \( c' \).
This leaves the question of whether it is computationally cheaper to update the upper and lower bounds than to calculate the point-center distances. Elkan shows that this is indeed the case because the bounds can be efficiently updated by using the information carried over from previous iterations and the center-center distances computed and cached at the beginning of each iteration. As an optimization, the upper and lower bounds are re-calculated when the upper bound becomes out of date, that is, when the upper bound u is bigger than both the lower bound l and the half distance from the assigned center to another center s. This tightening step can further eliminate some point-center distance computations.
In summary, Elkan results show that, in most cases, the upper and lower bounds are enough to determine whether a data point x will change its center assignment, so there is no need to compute the exact distance between x and a center c, and updating the upper and lower bounds does not necessarily require explicit calculation of point-center distances either. Hence, Elkan’s algorithm can significantly speed up the k-means clustering process by avoiding a large number of redundant point-center distance calculations.

Elkan’s algorithm is a very efficient method for k-means clustering, especially for high-dimensional datasets. The method does not use any indexing structure (e.g., a k-d tree) or pre-processing; instead, it keeps a number of distance bounds that allow it to avoid unnecessary distance computations. It is simple to implement and can provide dramatic speedups of up to 40x. However, storing k lower bounds for each data point, the algorithm keeps nk lower bounds in total, which consumes a substantial amount of memory if both k and n are large. Hamerly (2010) improves the algorithm in this regard, presenting an method that maintains only 1 lower bound l(x) per data point x. The upper bound in Hamerly’s algorithm is kept the same as in Elkan’s one, but the meaning of the lower bound is different. It now represents the distance to a data point’s second-closest center. That is, the 1 lower bound corresponds to the minimum distance that any center other than the data point’s currently assigned center can be to the data point.
If \( u(x) \le l(x) \), it is not possible for any center to be closer to x than the assigned center, which can be easily proved by applying Proposition 2 above. In this case, the computation of the distances between x and the k centers can be avoided, that is, the corresponding loop can be skipped as a whole. On the other hand, if \( l(x) < u(x) \), the closest center for x may change. In this case, the algorithm firstly tightens the upper bound by computing the exact distance d(x, c) and then checks the upper and lower bounds again to see if the \( u(x) \le l(x) \) condition is satisfied. If yes, it skips the distance calculations between data point x and the k centers. If not, then it goes into the innermost loop to compute those distances. The pseudo-code in Algorithm 2 shows the workflow of Hamerly’s algorithm.

Hamerly demonstrates that using only one lower bound per data point eliminates the innermost loop 80% of the time or more. Compared to Elkan’s algorithm, which has k lower bounds each data point, Hamerly’s algorithm has a much smaller memory overhead. However, this makes it less capable of reducing the required distance calculations. Hamerly’s algorithm works better than Elkan’s one with low-dimensional datasets only (Hamerly 2010) because centers tend to move more in high-dimensional datasets.
Drake and Hamerly (2012) aim to find a balance between memory footprint and the capability of pruning redundant distance calculations. Based on Elkan and Hamerly’s algorithms, they propose an accelerated method with adaptive distance bounds, by using \( 1< b < k \) lower bounds on the b closest centers for each point. The algorithm keeps the same upper bound on the distance to each point’s assigned center, but tracks b lower bounds per point on the distances to its b next-closest centers, always ordered by increasing distance, where \( 1< b < k \). The value of b can be selected in advance or adaptively learned while the algorithm runs. Thus, Drake and Hamerly’s adaptive method leverages the complementary strengths of Elkan’s and Hamerly’s algorithms by keeping a variable number of lower bounds. Experimentally, their work implies that for \( k > 8 \), k/8 is a good floor for b. For sufficiently large k, this hybrid approach achieves superior efficiency on medium-dimensional data. However, the mechanism required to maintain a variable number of bounds is more expensive than maintaining all k bounds per point as in Elkan’s method or simply 1 bound per point as in Hamerly’s approach.
There are other algorithms reported in the literature that are based on the triangle inequality, such as annular k-means (Hamerly and Drake 2015), which works similarly to Hamerly’s algorithm but with norm-ordered centers, and the heap k-means (Hamerly and Drake 2015), which uses k heaps of assigned points, ordered by bounded distance from the corresponding center, and combines the upper and lower bounds into one value.
As discussed above, using the triangle inequality to inexpensively maintain a set of distance bounds between points and centers is an idea with great benefit. Hamerly’s algorithm simplifies Elkan’s algorithm, so it greatly reduces the overhead of keeping bounds, but still allows the algorithm to skip the innermost loop across all centers in many cases. We base our GPU implementation on Hamerly’s approach because using less memory for the bounds becomes a big advantage for GPU computing. Storing more bounds not only takes more device memory, which is a precious resource in GPU computing, but also requires more time to transfer the data between the main memory and the device memory due to the increased size. It also takes more time for the GPU to write the data into the device memory (writing to the device memory is relatively slow). Crucially, similar to Elkan’s algorithm, Drake and Hamerly’s method is capable of partially skipping the loop that computes distances between a point and all the centers, but this does not reduce the number of data points that require transfer to GPU memory: even if the distance to only one center is required to be computed for a data point, it needs to be uploaded to the GPU.
2.4 GPU-based K-means implementations
GPUs can be used to significantly accelerate several clustering problems. Previous research shows that GPUs outperform CPUs and concludes that GPU-based implementations can solve complex clustering problems (Jian et al. 2013; Chiosa and Kolb 2011): Clustering can be sped up substantially by offloading work to a GPU. Here, we focus on k-means. The main idea of GPU-based k-means is that data-parallel, compute-intensive portions, e.g., data object assignment, of traditional k-means can be off-loaded from the host to the device to improve performance.
Lee and Chu (2012) implement the hierarchical k-means algorithm using CUDA, with nearest centroid finding and centroid updating running on the GPU. To achieve a higher level of coalesced data access, a data rearrangement process is introduced, which is executed before initialising the next layer.Footnote 1 The GPU implementation achieves 700x speedup over the CPU version on an artificial dataset, randomly generated, with 128 dimensions.
Farivar et al. (2008) speed up k-means clustering by running the data point assignment (labelling) step on a GPU. One important aspect of the implementation is the use of device constant memory (i.e., read-only memory) for storage of the centroid data. Their CUDA implementation guarantees the same output as the original CPU-based k-means algorithm and achieves an over 13x performance improvement compared to a baseline 3 GHz Intel Pentium(R) based PC when running the same algorithm with a G80 graphics card, the NVIDIA 8600GT. However, the use of constant memory for centroids may not be practical when the number of centroids is large and the dimensionality is high. NVIDIA GPUs usually provide 64KB of constant memory (or less). For a dataset with 50 dimensions, only 320 centers can be loaded into it.
Zechner and Granitzer (2009) present an optimized k-means implementation on the GPU. The algorithm performs distance calculations on the GPU in a parallel fashion while sequentially updating cluster centroids on the CPU. They overcome some drawbacks and limitations of previous related work, e.g., maximum data size and clusters, by storing data points and centroids in global memory. A transposed matrix is used to represent data points in global memory to favour coalesced access. Centroids are loaded into shared memory in a batch fashion in each thread block to reduce memory access latency. An empirical performance study on synthetic data using an NVIDIA GeForce 9600 GT demonstrates a maximum 14x speed increase compared to a fully SIMD optimized CPU implementation. As the authors point out, the implementation is not suitable for sparse data in a high-dimensional space, e.g., document collections, and the GPU’s computational power is not fully utilised due to the memory-bound nature of the implementation.
Hong-Tao et al. (2009) accelerate k-means by off-loading both the data objects’ assignment and the recalculation of the k centroids onto the GPU. Multiple threads are employed to avoid memory access latency in the assignment stage. In order to update the centroids efficiently, the cluster memberships are downloaded from the GPU, all data points are rearranged, and the number of data objects contained in each cluster is counted on the CPU. Then this information is uploaded to the GPU to get the centroids updated. Data points and centroids are stored in global memory because the constant and shared memory are limited in size. Their experiments show that the GPU-based k-means runs 8 to 14 times faster than its CPU-based counterpart when running on a PC with an Intel Pentium D 965 CPU 3.7 GHz with 1GB main memory and an NVIDIA GeForce 8800 GTX graphic card with 768MB device memory.
Jian et al. (2013) propose three CUDA-based parallel techniques: (1) a scalable thread scheduling scheme for irregular patterns; (2) a parallel distributed top-k scheme; and (3) a parallel high dimension reduction scheme. Then, they present a parallel implementation of the k-means algorithm by off-loading the whole iterative process onto the GPU, that is, the GPU performs data point assignment, centroid recalculation, and centroid movement detection. The latter two are implemented using their parallel high dimension reduction scheme. The scalable thread scheduling scheme is also applied in the k-means implementation. The experimental results obtained on the KDD-CUP 1999 intrusion detection dataset show that 2 to 5 times speedup is observed when compared with the other state-of-the-art implementations (Fang et al. 2008).
Lutz et al. (2018) presented an approach that performs center assignment and update steps of an iteration with a single pass over the data points, which all have to be loaded into the GPU’s global memory. A couple of optimizations have been explored in order to improve the throughput of the GPU, including a GPU-optimized center update algorithm and a single-pass execution strategy. By applying their approach on subsets of the data at a time, the algorithm is able to handle large datasets. However, the whole dataset still needs to be loaded onto the GPU multiple times, once for each iteration, which is inefficient. The algorithm is also incapable of efficiently handling high dimensional datasets with a large number of centers due to the limited GPU local memory, which is needed for some essential data manipulation operations.
Yang et al. (2020) examined the sensitivity of two CUDA implementations of k-means to the number of clusters, dataset size and data dimension. One implementation uses the GPU’s global memory to store cluster centers, the other uses shared memory. All results were obtained by loading the whole dataset into the device’s global memory. No attempt was made to address the efficiency of transferring large datasets multiple times between the CPU and GPU.
Although many papers have been published on accelerating k-means using GPUs, almost all of them are based on the standard k-means algorithm by applying different optimization techniques on various steps and require the whole dataset to be loaded into the GPU’s global memory (Kruliš and Kratochvíl 2020; He et al. 2022; Taylor and Gowanlock 2021). To the best of our knowledge, massively parallel processing of the k-means clustering algorithm accelerated with the triangle inequality has not been reported in the literature, especially an algorithm that is capable of handling datasets larger than the global memory of the GPU.
3 Proposed algorithm
Our proposed GPU-accelerated algorithm is capable of handling big datasets efficiently by combining elements of Hamerly’s k-means, batched processing, and GPU acceleration for massively parallel processing. As reviewed above, Hamerly’s algorithm is a simplification and modification of Elkan’s k-means algorithm. Unlike Elkan’s algorithm, which uses k lower bounds for each data point, Hamerly’s algorithm uses 1 lower bound per data point. This greatly reduces the memory usage, simplifies the logic, and also decreases the amount of data required to be transfered between the device and CPU.
We split the clustering method into three components: The data point selection step, the membership assignment step, and the center and bound update step. The data point selection and center update steps are performed on the CPU (asynchronously) while the assignment and bound update steps are performed on the GPU. With sufficient statistics maintained in main memory, the center update step is very efficient and takes little time to finish. The mechanism used in Hamerly’s algorithm to select data points for the next iteration is applied in the data point selection step. Similar to Hamerly’s algorithm, our algorithm uses efficiently updated distance bounds and the triangle inequality to select those points that may change their cluster membership in the next iteration. The point-center distance needs to be re-computed for these data points only. In summary, the proposed algorithm has the following features:
-
1.
It uses the triangle inequality to select the data points that need to be processed in each iteration (based on the selection criteria used in Hamerly’s accelerated k-means). These data points require computing the distances to all the centroids, which is the most compute-intensive part of the algorithm. Data point selection not only decreases the time spent on distance computations but also saves time on data transferal.
-
2.
The data point selection and center update steps can be performed asynchronously on the CPU while the GPU is computing point-center distances for a selected batch of data points. In most existing GPU k-means implementations, the CPU is idle when the GPU is performing arithmetic computations. Consequently, most of CPU’s computing power is wasted.
-
3.
The algorithm makes use of the strengths of both the CPU and the GPU: The CPU performs the logical checks (in the selection step) and sequential processing, while the GPU performs floating-point arithmetic operations and massively parallel processing.
-
4.
The amount of data transferred between the CPU and the GPU is comparatively small. In most cases, the number of data points that require re-computation of the distances to all centroids is relatively small, which is especially true in the later iterations of k-means. Most data points will not change their memberships often between iterations. Based on our experiments, in most cases, 90% or more of data points do not change their cluster assignment after the first few iterations.
-
5.
The algorithm generates the same output as the standard k-means algorithm, and it is also possible to use distance functions other than the Euclidian distance in the membership assignment step running on the GPU.
As shown in the experiments, compared with Elkan and Hamerly’s k-means algorithms, which are two of the fastest implementations known for CPU-based k-means clustering, our GPU-based implementation speeds up the clustering process significantly, while still obtaining the exactly same clustering results.
3.1 GPU-based triangle-inequality-accelerated k-means clustering
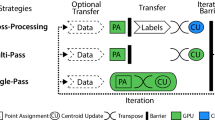
To facilitate the understanding of the algorithm, we describe a simplified version with synchronized data transfer first. As shown in Figure 1, the algorithm works as follows.
-
1.
Create the initial set of centers: the k-means++ initialization algorithm (Vassilvitskii and Arthur 2006) is used for this. k-means++ provides statistical guarantees on the quality of clusterings. The upper and lower bounds and the assignments are also initialized. This step is performed on the CPU.
-
2.
Select a batch of data points: the triangle inequality is used to select the data points, which decreases the amount of data transferred to the device and avoids redundant distance computations. This is realized by using 1 upper bound and 1 lower bound per data point—the mechanism used in Hamerly’s k-means algorithm. During data selection, the data representation is also converted from a row-major matrix to column-major to facilitate coalesced memory access in the global memory on the GPU. The selected batch of data is then transferred to the device. This step is performed on the CPU.
-
3.
Compute the point-center distances: The GPU computes the distance to all centers for each data point and finds out their cluster assignments. The distance to the closest and second closest centers are also recorded. These distance computations are expensive for the standard CPU-based k-means algorithm. They are delegated to the GPU, which is more suitable for performing arithmetic operations. The cluster assignments, together with the two distances, are transferred back to the main memory after the GPU finishes computations.
-
4.
Update the sufficient statistics: A data structure is constructed in main memory to capture the information required for center and bound updates, including 1) the sum and count of the data points in each cluster, 2) the cluster membership assignments, and 3) the distance to the closest and second closest centers. This information is efficiently updated by the CPU after each batch of data is processed by the device.
-
5.
Repeat step 2-4 until all data points have been processed.
-
6.
Update the centers: This step is performed very efficiently by the CPU based on the sufficient statistics cached in main memory. New centers and their movements are calculated. The two biggest movements are used to update bounds in the next step of the algorithm, as discussed in Section 3.6. This information, together with the cluster membership assignments and the upper and lower bounds recorded in Step 4, are then uploaded to the device.
-
7.
Update bounds and inter-center distances: The upper and lower bounds are updated on the GPU, and the inter-center distances are computed. Then, the updated bounds and inter-center distances are downloaded to main memory so that they can be used to select data points for the next iteration.
-
8.
Repeat Step 2-7 until the centers converge.
In the synchronized implementation, all steps are executed sequentially, that is, no operation is performed simultaneously on the CPU and GPU. When one is busy performing a task, the other is idle waiting. The computational powers of the CPU and GPU are not fully used.
Figure 2 shows a detailed description of the asynchronous version of the algorithm, which involves the same steps as in the synchronized version, except that 1) the data are transferred asynchronously to/from the device, and 2) some tasks are performed simultaneously on both the CPU and the GPU. For each batch in each iteration, the CPU selects data points for the next batch and updates sufficient statistics using results from the previous batch, while the GPU is performing the center-point distance computations. A small optimization is introduced for the creation of the first batch of data in an iteration. The first batch is created on the CPU while the GPU is updating the upper and lower bounds and computing the inter-center distances. At this point in time, because there is no updated information for data selection using the triangle inequality, we simply select the first n data points from the dataset, and convert the data from a row-major matrix to a column-major representation.
Because of the batched processing, the algorithm can handle datasets that are bigger than the size of the global memory on the device. In theory, one can choose to load data from a fast mass storage or data stream. In our implementation, we only handle datasets that can be loaded into the computer’s main memory. (Note that this is generally much larger than the GPU’s global memory.)
Some of the other algorithm details, such as the asynchronous data transfer, distance computations on the GPU, update of sufficient statistics, and update of bounds, etc., are described below.
3.2 Data representation on the GPU
In our implementation, we use the GPU’s global memory for storage of the centers and batch of data points. Reading data from global memory is slower than from shared or constant memory, but if there are sufficient math instructions in the kernel and enough threads to hide the latency, the global load/store instructions are a minor cost. A possible optimization is to load centers in the device’s shared memory tile-by-tile (Li et al. 2013). However, based on our experiments, using shared memory only slightly improves the performance on modern GPUs. Moreover, the shared memory is limited in size and using shared memory for a large number of centers greatly increases the complexity of the code.
Appropriate layout of data is crucial for maximum performance on the GPU. To facilitate coalesced global memory access, data instances are stored on the GPU in a column-majored matrix with each column representing an instance. Each row of the data matrix stores values of a dimension. Centers are stored in a matrix on the GPU with each row representing a center.
To facilitate computation of the upper and lower bounds, in addition to saving the cluster assignments, i.e., the index of the cluster each instance belongs to, we also need to save the distances to the first two closest centers to implement the algorithm discussed above. So, the data matrix on the GPU has three extra rows: the cluster assignment is stored in the first row, the distance to the closest center in the second row, and the distance to the second-closest center in the third row.
The GPU is responsible for finding the cluster an instance belongs to by computing the distance to every center. The distances to the closest and the second-closest centers are recorded. Algorithm 3 describes the GPU kernel for the distance computation. Each GPU thread processes one instance, computing the distance between the point and each center. While executing this loop, it also tracks the minimum distance to the instance’s nearest center, and when the loop is completed, the results will be stored in the device memory. Because this information is required for updating the upper and lower bounds, it is transferred back to the host together with the assignments after the GPU finishes processing all the data points.

3.3 Data transmission
The main goal of optimizing host-device interaction is to minimise the time spent on data transferal. This can be achieved directly by minimising the amount of data transferred between the host and the device and indirectly by overlapping GPU kernel execution with memory copies. The centers are uploaded to the GPU’s global memory before an iteration starts, which happens once only per iteration. The size of the centers is relatively small compared to the size of the whole dataset, so the time taken for transferring centers is negligible. However, data points need to be uploaded to the GPU in each iteration in a batched fashion. Although the data selection mechanism significantly reduces the number of data points that require uploading, transferring this data still consumes some time. This data transfer should be performed asynchronously, i.e., the data for the next batch is uploaded to the GPU while the previous batch is being processed on the GPU.
Asynchronous operation means the host enqueues work and returns immediately without waiting for the completion of the work. Unlike GPU kernel launches, which are inherently asynchronous and automatically overlap with host operations, transferring data asynchronously requires special treatment, involving the use of CUDA constructs specifically designed for the purpose.
In CUDA, streams and events are the enablers of asynchronous operations. A stream is a queue of device work. The host places work in the queue and continues immediately. The device schedules work from streams when resources are free. CUDA operations are placed within a stream, e.g., kernel launches and memory copies, etc. Operations within the same stream are ordered and executed in a FIFO fashion and they cannot overlap. Operations in different streams are unordered and can overlap. A race condition may occur between different streams so care is required when using these facilities.
In order to achieve overlapped data transfer over compute, the following requirements need to be satisfied (NVIDIA 2021):
-
1.
Transfers must be in a non-default stream because the default stream is a synchronizing stream.
-
2.
Async copying APIs must be used.
-
3.
Perform one transfer per direction at a time. That is, there is not another memory copy occurring in the same direction at the same time.
-
4.
Memory on the host must be pinned.
Asynchronous data transfer requires the use of pinned (i.e., page-locked) host memory, which is allocated using special allocators and cannot be paged out by the OS. Pinned memory is transferred using the system’s DMA engines instead of using the host CPU as in data copying using pageable memory. This frees the CPU for asynchronous execution and achieves a higher percentage of peak bandwidth.
CUDA events can be used to synchronize some operations between the host and the device. A CUDA event is an object with one of two Boolean states: "occurred" (the default state) or "not occurred". It provides a mechanism to signal when operations have occurred in a stream, and is useful for profiling and synchronization. In our implementation, CUDA events are used to synchronise device operations with the host and between streams when a synchronisation is required.
To achieve asynchronous data transfer, two blocks of global memory are allocated on the device: one block stores the data being processed and the other receives the data for the next batch. A GPU stream is used for this data transfer. Since pinned memory required for asynchronous data copying is a limited resource, it should be used wisely—otherwise the system’s performance may be negatively affected. In our implementation, a fixed, small size of pinned memory is allocated and reused. Multiple asynchronous copying operations may be required when the size of the data for a batch exceeds the amount of pinned memory allocated. After a batch of data has been processed on the device, the results are copied back to the main memory asynchronously using another GPU stream.
3.4 Sufficient statistics
In order to update the centers and bounds efficiently on the CPU, we need to cache sufficient statistics in main memory. For each cluster, we maintain a vector sum of the points assigned to the cluster and the number of points assigned to the cluster. Keeping this information is inexpensive and avoids a sum over all points for each iteration. Each time a point changes cluster membership, the relevant sufficient statistics are updated. After the first few iterations, most points remain in the same cluster for many iterations. Thus, updating sufficient statistics is much cheaper than a sum over all points. For each data point, we maintain two values in memory, one for the upper bound and the other for the lower bound. We also need to maintain an array to store the cluster membership assignment of each data point.
3.5 Data point selection
As described in Algorithm 4, we go through the whole dataset to select the data points for which the membership assignment may change and require re-calculation of the distance to all centroids. The selection process is based on the lower and upper bounds and the inter-center distances. A data point will not be selected if it satisfies the following conditions, because its membership will not change in the next iteration:
-
For a data point i, if its upper bound \( u \left( i\right) \) is less than or equal to its lower bound \( l \left( i \right) \), the centroid assignment of the data point will not change in the next iteration.
-
The data point will not change its centroid if the upper bound \( u \left( i\right) \) is less than or equal to the distance to its second closest centroid \( s \left( c \left( i \right) \right) \) divided by 2.
In summary, if the condition \( u \left( i \right) \le \max \left( l \left( i \right) , s \left( c \left( i \right) \right) / 2 \right) ) \) is true, the data point will not change its centroid assignment in the next iteration, so it will not be selected. If a data point is selected, it is added into the buffer for the next iteration, and the index to the original position in the data set is recorded. When the maximum number of data points have been selected, the selection process terminates and the index of the last processed data point is recorded. The selection process for the next batch begins from this point.

3.6 Center update
With sufficient statistics maintained in the main memory, center updates can be performed efficiently on the CPU. Center movements are also computed in this step and the largest and the second-largest center movements are recorded for updating the lower bound later.
Algorithm 5 describes the center update process. It also finds the index of the center that moves the most, \( \gamma \), and the index of the second-largest mover \( \delta \).
Note that if the number of centroids is large (for example, we may have thousands or tens of thousands of centers in some image segmentation applications), it may be useful to move the centroid update to the GPU, which can reduce the time consumed, but this is not currently done in our implementation.

3.7 Updating the bounds
The upper or lower bound for each data point can be updated efficiently by adding or subtracting the distance moved by a center each time when the center moves after an iteration. This allows us to maintain both upper and lower distance bounds between a point and a moving center without explicitly calculating distances. Updating bounds for every data point requires a loop of the size of the dataset. Although this step can be performed efficiently on the CPU, due to the large size of the datasets considered, performing the task on the GPU is much faster, with one GPU thread updating the bounds for one data point.
When the step is performed on the GPU, the center movements, the index to the largest and the second-largest center movements obtained in Algorithm 5, together with cluster membership assignments, and the upper and lower bounds, are transferred to the global memory asynchronously. Algorithm 6 describes the process on the GPU.

3.8 Inter-center distance computations
Our algorithm also requires a function to compute the inter-center distances, keeping only the closest distances, and updating s. After this, \( s \left( c \right) \) will contain the distance between the center c and its closest other center, divided by two. The division here saves repeated work later, since the algorithm always requires the distance divided by 2. This step can be performed efficiently on the CPU when the number of centers is small. For a large number of centers, it is much cheaper to execute it on the GPU. Because we have downloaded the required data to the global memory in Algorithm 6 above, it is trivial to calculate the smallest inter-center distance on the GPU. Algorithm 7 describes the process.

4 Experimental results
We now proceed to present empirical results obtained using our algorithm, for two real-world and one synthetic domain, comparing to relevant baselines.
4.1 Experimental configuration
The computing environment used to execute and test our algorithm is equipped with a CPU Intel Core i7 (2.8 GHz), 64GB of RAM and an NVIDIA Titan X GPU, with 3584 CUDA cores, 11 Tera ops in single precision (i.e., 11 TFLOPS), 12GB GDDR5X of device memory size, and 480GB/s of memory bandwidth. The operating system is Ubuntu 16.04 server edition with the CUDA version 10.2 GPU computing platform installed.
4.2 Datasets
In our experiments, three different types of datasets are used:
-
1.
datasets containing large collections of image patches
-
2.
Twitter Glove datasets
-
3.
synthetic datasets
The image datasets are composed of small grayscale image patches, which are collected from unlabelled imagery by cropping out random n-by-n chunks. Each n-by-n pixel grayscale patch is represented as a vector of \(n \times n\) pixel intensities (i.e., \( x(i) \in R^{(n \times n)} \)). The method described in the paper (Coates and Ng 2012) is used to normalize the brightness and contrast of the patches: for each x(i) , we subtract out the mean of the intensities and divide by the standard deviation. A small value is added to the variance before division to avoid dividing by zero and also suppress noise. The images used to create the image patches datasets are obtained from Google image collections that are used to train the TensorFlow deep learning platform for animal species recognition.
The Twitter Glove dataset (Glove.Twitter.27B) is downloaded from the Global Vectors for Word Representation project (Pennington et al. 2021) established by the Natural Language Processing Group at Stanford University. It is a dataset containing pre-trained word vectors generated from a word corpus of 2 billion tweets, which contains 27 billion tokens and 1.2 million vocabularies. The dataset contains word vectors with 50, 100 and 200 dimensions, respectively. The 200d word vector dataset is used in this paper to demonstrate that our algorithm can cope well with high-dimensional real-world datasets.
The synthetic datasets are created by using a random data generator producing a uniform data distribution, so that no natural clustering structure exists. This is to avoid biases introduced by pre-clustered data and represents the worst-case scenario for any clustering algorithm.
Note that the size of some datasets used in the experiments is smaller than the total amount of device memory available on the GPU so that the whole dataset can be loaded into the GPU global memory. This is because some algorithms used in our experimental comparison require the whole dataset to reside in the GPU’s memory. However, we also present results obtained on datasets that are bigger than the GPU’s global memory, in order to demonstrate that our algorithm is capable of also handling such large data with good performance.
4.3 Initialization
To avoid some common problems like empty clusters, Lloyd’s algorithm for k-means clustering algorithm is known to require a number of small tweaks (Bahmani et al. 2012). One important consideration is the choice of initial centers. Though it is common to solve this problem by randomly choosing instances from the data to be centers, this does not work well in practice. Data points may tend to group too densely in some areas, and thus initializing k-means with randomly chosen vectors may lead to a large number of centers starting close together. Many of these centers ultimately end up becoming near-empty clusters. Another way to pick initial centers is k-means++ (Vassilvitskii and Arthur 2006), which we apply in our experiments. The idea is to still randomly choose instances, but with a probability which is not uniform: it is proportional to the distance of each remaining instance to the nearest center selected so far. K-means++ ensures that the centroids selected in the initialization stage are as far as possible. From a theoretical side, the k-means++ refinement is proven to make Lloyd’s process reach the optimal solution with an approximation ratio \( O(\log k) \) in expectation.
4.4 Batch size
In order to understand how different batch sizes affect the runtime of our approach, we measure the runtime of the algorithm on different datasets using different batch sizes. Figure 3 and 4 shows the impact of different batch sizes on execution time. The batch sizes are multiples of the number of the GPU cores. 1x means the number of instances in a batch is the same as the number of GPU cores, 2x means the number of instances in a batch is 2 times of the number of GPU cores, and so on. No obvious change to the execution time is observed for batch sizes from 1x up to 10x. The execution time increases slightly with batch sizes from 20x to 200x, and increases dramatically when the batch size is 500x and 1000x. When the batch size is big, the time spent on selecting a batch of instances on the CPU is longer than the time spent on clustering a batch of instance on the GPU, and this causes the GPU to idle and wait. In addition, big batch sizes decrease the parallelism of the algorithm, especially in the later iterations where the number of instances that will be selected for distance calculation is small.
The same pattern on the execution time is also observed on other datasets used in our experiments. Based on these results, in the further experiments presented below, a batch size of 35,840 data points is used, which is 10 times the number of GPU cores.
4.5 Performance evaluation
In this section, we present the results of running our algorithm on different datasets and compare them to the results generated by other k-means algorithms on the same datasets. Firstly, we compare our algorithm with the k-means implementation from the NVIDIA RAPIDS library. Then, we look at the performance of our algorithm in terms of execution time together with a GPU version of the Lloyd k-means algorithm, and CPU versions of Elkan’s and Hamerly’s accelerated k-means algorithms. In order to make the results comparable, the data used for comparison were all generated based on the same experimental configuration, that is, the same initial set of centers is used by all algorithms in an experiment, and the number of iterations is fixed to 300 in all experiments.
4.6 ASB k-means vs. RAPIDS k-means
RAPIDS k-means is the k-means implementation from the RAPIDS Machine Learning Library (cuML). The RAPIDS suite, created by NVIDIA, is a suite of open-source software libraries aiming to enable execution of end-to-end data science and analytics pipelines entirely on GPUs. It relies on NVIDIA CUDA primitives for low-level compute optimization, but exposes that GPU parallelism and high-bandwidth memory speed through Python interfaces. cuML, included in the RAPIDS suite and sharing compatible APIs with other RAPIDS projects, is a set of libraries that implement machine learning algorithms and mathematical primitive functions. NVIDIA claims that for large datasets these GPU-based implementations can complete 10-50x faster than their CPU equivalents.
Firstly, we compare the performance of our ABS-KM algorithm and the RAPIDS-KM on synthetic datasets. Table 1 lists all the datasets used in the experiment.
4.6.1 Results for synthetic datasets
Figure 5 shows that ABS-KM is consistently faster than RAPIDS-KM on all datasets. From Table 2, we can see that ABS-KM is slightly (i.e., 1.2 times) faster than RAPIDS-KM on the synthetic dataset with 300K instances, where ABS-KM and RAPIDS-KM take 43.78 and 52.46 seconds, respectively. ABS-KM performs about 1.5 times faster than RAPIDS-KM on all other datasets.
4.6.2 Results for the image datasets
Figure 6 shows the execution time of ABS-KM and RAPIDS-KM running on different image datasets. On all these datasets, ABS-KM outperforms RAPIDS-KM. For the dataset with 1.5 million instances, ABS-KM uses about half of the time taken to run RAPIDS-KM. Similar results are also observed on the datasets with 900K and 1.2M instances.
4.6.3 Results for the twitter datasets
ABS-KM performs slightly better than RAPIDS-KM on the three Twitter datasets, as illustrated in Figure 7. ABS-KM runs 40% faster on the Glove.Twitter.27B.50D dataset and 15% faster on the Glove.Twitter.27B.100D dataset, and marginally outperforms RAPIDS-KM on the Glove.Twitter.27B.200D dataset.
4.6.4 Execution time using different numbers of centers
To understand how ABS-KM performs when the number of centers increases, we evaluate both ABS-KM and RAPIDS-KM on the synthetic dataset with 1.5M instances of 100 dimensions, the image data with 900K instances and 256 dimensions, and the Glove.Twitter.27B.50D dataset, with the number of centers varying from 100 up to 1500. The results are shown in Figure 8, 9 and 10. ABS-KM out-performs RAPIDS-KM in all the experiments.
4.7 ASB k-means vs. Elkan k-means, Hamerly k-means, and Lloyd k-means
The k-means algorithms used in this experiment are listed below:
-
1.
Asynchronous Selective Batched K-means (ASB-KM)
-
2.
GPU-based Lloyd/naive k-means (NK-GPU)
-
3.
Elkan k-means running on CPU with 8 threads (Elkan-KM-8T)
-
4.
Elkan k-means running on CPU with 1 thread (Elkan-KM-1T)
-
5.
Hamerly k-means running on CPU with 1 thread (Hamerly-KM-1T)
-
6.
Lloyd k-means running on CPU with 1 thread (Lloyd-KM-1T)
4.7.1 Synthetic datasets
Seven synthetic datasets were used in our experiment, as shown in Table 8. The smallest dataset, which is 400MB in size, contains 200 thousand instances with 500 attributes each, while the largest one contains 2 million data points with 512 attributes each, and is 4,096MB in size.
Firstly, we consider how different k-means algorithms perform on rnd-ds1 and rnd-ds2. Figure 11 shows the execution time of ABS-KM, NK-GPU, Elkan-KM-8T, Elkan-KM-1T, Hamerly-KM-1T and Lloyd-KM-1T on synthetic dataset 1 and 2 with 256 centers. On dataset 1, Hamerly’s algorithm is 4.31 times faster than GPU-based Lloyd (naive) k-means, Elkan k-means is 21.83 times faster when 1 CPU thread is used and 99.27 times faster when 8 CPU threads are used. The 8-thread version of Elkan k-means out-performs the GPU-based Lloyd (naive) k-means, which is 86.11 times faster than the CPU-based implementation of Lloyd’s algorithm. Our k-means implementation (i.e., ASB-KM) is the most efficient one, being 143.63 times faster than the CPU-based Lloyd implementation. The same trend can be seen on the synthetic dataset 2 as demonstrated by the results shown in Table 9. Since Lloyd’s and Hamerly’s k-means took much longer to finish than Elkan’s k-means on all the datasets used in our experiment, we just show the results of these two slower algorithms on the two synthetic datasets. In our experiments, among the three CPU based k-means implementations, Elkan’s algorithm always clearly outperforms the other two. Therefore, amongst the CPU-based algorithms, only Elkan’s algorithm is used for comparison in the remaining experiments.
Table 10 shows the execution time of ASB-KM, NK-GPU, Elkan-KM-8T, and Elkan-KM-1T on different synthetic datasets, and the results is also graphically displayed in Figure 12. The single-thread Elkan algorithm ran the slowest on all datasets. It is used as a baseline here. On the three smaller datasets, the eight-threaded Elkan algorithm ran slightly faster than the GPU-based Lloyd k-means (i.e., NK-GPU) implementation, and NK-GPU outperformed Elkan-KM-8T on the two larger datasets (rnd-ds4 and rnd-ds5). Our ASB-KM algorithm performed the best on all the datasets, and is 6.58x, 6.30x, 11.76x, 8.27x and 9.14x faster than Elkan-KM-1T respectively.
Secondly, we investigate how the three k-means implementations perform with different numbers of centers. Synthetic dataset 6 was used in this experiment. The results are shown in Table 11 and Figure 13. It can be observed that ASB-KM and Elkan-KM-8T outperformed NK-GPU in all experiments on this dataset with different numbers of centers. Elkan-KM-8T performed the best, and is 1.56 times faster than NK-GPU, when the number of centers is small, i.e., 200. Our ASB-KM is just slightly faster than NK-GPU in this case. However, ASB-KM performs better than Elkan-KM-8T when the number of centers is equal to or larger than 400.
Lastly, we look at how well our ASB-KM is able to handle a large number of centers. Table 12 and Figure 14 show the runtime of ASB-KM k-means algorithm on the synthetic dataset 7 with different numbers of centers. The results show that our algorithm scales well with the number of centers. The time taken by the algorithm using 50 centers is 2850.67 seconds and it is 4098.12 seconds for 1500 centers: the number of centers increased by a factor of 30 while the time taken just increased by a factor of 1.4.
4.7.2 The image datasets
Table 13 lists the five image datasets used in the experiments, which were created from Google’s image collection used for animal and plant species classification and recognition as described in the dataset generation section above. The smallest dataset is 2048 MB in size and contains 2 million of instances with 256 features each, while the largest one has 60 million of instances with 256 features each, and the total size is 61,440MB.
Table 14 and Figure 15 show the runtime of ABS-KM, NK-GPU, Elkan-KM-8T and Elkan-KM-1T on the image datasets 1 and 2. Dataset 1 & 2 are used here because they can be loaded into the GPU’s global memory as a whole, which is required by the GPU-based Lloyd k-means implementation (i.e., NK-GPU).
It took 527.70 seconds for Elkan-KM-1t to complete the clustering process on the image dataset 1, which is the slowest in the four implementations, while it took 217.24 seconds for NK-GPU, 125.81 seconds for ASB-KM, and 122.63 seconds for Elkan-KM-8T. ASB-KM is very close to Elkan-KM-8T performance wise on this dataset, which is the smallest in the five datasets. On image dataset 2, it took 547.63, 742.23, 1168.33 and 3034.78 seconds for ASB-KM, Elkan-KM-8T, NK-GP and Elkan-KM-1T respectively to complete the clustering process, where ASB-KM, Elkan-KM-8T and NK-GPU are respectively 5.54x, 4.09x and 2.60x times faster than Elkan-KM-1T. ASB-KM outperforms the other three implementations on the larger of the two datasets.
To understand how our algorithm scales with respect to the number of data points, we perform the same tests discussed above with datasets larger than the size of the GPU’s global memory. In general, the previously described trends hold for the larger data sets as can be seen in Table 15 and Figure 16. For example, it took 7235.92 seconds for ASB k-means algorithm to cluster Image Dataset 5, whereas it would take days for other CPU-based algorithms to complete the clustering process.
Table 16 and Figure 17 show the time taken by ASB k-means to cluster the Image Dataset 1 with different numbers of centers. It took 36.76 seconds to cluster the dataset into 50 groups (i.e., centers), and 87.12, 194.67, 632.75 and 1374.81 seconds for 100, 200, 500 and 1000 centers respectively. Hence, the algorithm performed well when the number of centers/clusters increased by 20 times.
4.7.3 The twitter datasets
Table 17 shows the number of instances, dimensions, and size of the three Twitter datasets used in the experiments. All three datasets contain the same number of instances with different numbers of dimensions, that is, 50, 100 and 200 respectively.
For each dataset, the execution times of different k-means algorithms are obtained using 100, 500 and 1000 centers. Table 18 and Figure 18 display the runtime of different algorithms on the three datasets with 100 centers. It can be seen that ASB-KM completed the clustering process in 17.09 seconds on the ds1 dataset, while the runtimes are 33.83, 55.5 and 230.81 seconds for NK-GPU, Elkan-KM-8T and Elkan-KM-1T respectively. ASB-KM performed the best, and is 13.51 times faster than Elkan-KM-1T, compared to 6.82 and 4.16 times for NK-GPU and Elkan-KM-8T respectively. Similar results were also observed on the ds2 and ds3 datasets. ASB-KM is 9.21 times faster than Elkan-KM-1T on the ds2 dataset, compared to 6.50 and 5.10 times for NK-GPU and Elkan-KM-8T respectively. Finally, ASB-KM is 10.04 times faster than Elkan-KM-1T on the ds3 dataset, compared to 6.51 and 5.79 times for NK-GPU and Elkan-KM-8T respectively.
Table 19 and Figure 19 show the runtime of different algorithms on the three datasets with 500 centers. Similar to the results on these datasets with 100 centers, ASB-KM outperformed all other algorithms in this experiment. It is 17.39, 12.22 and 10.12 times faster than Elkan-KM-1T on ds1, ds2 and ds3 respectively, followed by NK-GPU, which is 7.13, 5.33 and 4.40 times faster than Elkan-KM-1T. NK-GPU performed better than Elkan-KM-8T on the ds1 and ds2 datasets (7.13x vs 3.95x on ds1 and 5.33x vs 4.80x on ds2) but was beaten by Elkan-KM-8T on the ds3 dataset (4.40x vs. 5.49x).
Table 20 and Figure 20 show the runtime of different algorithms on the three datasets with 1000 centers. As expected, ASB-KM outperformed other algorithms on all the three datasets, with 14.13 times faster than the Elkan-KM-1T on the ds1 dataset, 10.61x on the ds2 dataset and 8.92x on the ds3 dataset.
Table 21 and Figure 21 summarize the runtime of the ASB-KM algorithm on the three Twitter datasets with different number of centers. It can be seen that the algorithm scales well with the number of centers. On all the three datasets, when the number of centers increased 10 times from 100 to 1000, the runtime increased by 8.11 times for the ds1 dataset, 5.89x and 9.21x for the ds2 and ds3 datasets respectively.
4.8 GPU memory usage of different k-means algorithms
One advantage of the ABS k-means algorithm is the small memory usage. Unlike most GPU-based k-means algorithms that load the whole dataset onto the GPU, the size of memory usage in ABS-KM is just a fraction of the whole dataset size: it is the size of the data points in one batch (i.e., batch size) plus the size of the centers. Table 22 shows the memory usage of three GPU-based k-means methods, observed during the experiments on the three Twitter datasets. It is obvious that the GPU memory usage of ASB-KM is small compared to the size of the whole dataset, especially when the size of the dataset is big. For example, ABS-KM uses 195 MB of GPU memory only when the size of the dataset is about 1GB, whereas memory usage is 1,055MB for Lloyd-KM and 4,719MB for RAPID-KM. ASB-KM’s low memory usage might make it feasible to cluster large datasets on edge devices with small memory GPUs, like mobile phones and smart sensors. This may, in some cases, eliminate the need of passing large amounts of raw data back to a powerful central server to analyse: instead a summary or excerpt of the data can be generated on the edge device by applying our algorithm and only this summary is then transferred to the central computing unit for further processing.
5 Conclusions and future work
In this paper, we have investigated the potential of applying the triangle inequality in a GPU-accelerated k-means implementation and proposed a new algorithm that can be used as a drop-in replacement of the classic k-means algorithm. Experiments on a variety of test datasets show significant speedup and demonstrate good scalability in handling large datasets with high dimensions, while consuming very limited GPU memory. Our GPU implementation achieves good results on both synthetic and real-world datasets when compared to other popular k-means implementations. The algorithm performs particularly well for large high-dimensional datasets that need to be clustered into many clusters. Using the triangle inequality, many distance re-computations can be avoided, which dramatically reduces the data transfer rates between CPU and GPU memory. These transfers are a major bottleneck for any GPU implementation that needs to split large data into batches that fit into GPU memory.
Considering future work, there are a number of GPU programming options that can be explored. These include loading cluster centers into shared GPU memory to improve access speed, general loop unrolling, and considering smaller block sizes, which may help to improve hardware load balancing. In addition, more of the processing could be moved onto the GPU. Currently only the most computationally expensive parts of the algorithm, which are cluster assignment and re-computation of the bounds, are executed on the GPU.
Notes
No detail is provided on how this process is implemented in Lee and Chu (2012).
References
Bahmani B, Moseley B, Vattani A et al (2012) Scalable k-means++. Proc VLDB Endow 5(7):622–633
Bejarano J, Koushiki B, Brannan T, et al (2011) Sampling within k-means algorithm to cluster large datasets. Tech. Rep. HPCF-2011-12, UMBC High Performance Computing Facility, University of Maryland, Baltimore County, Maryland, USA
Berkhin P (2006) A survey of clustering data mining techniques. In: Grouping Multidimensional Data. Springer, p 25–71
Brodtkorb AR, Hagen TR, Sætra ML (2013) Graphics processing unit (GPU) programming strategies and trends in GPU computing. J Parallel Distrib Comput 73(1):4–13
Che S, Boyer M, Meng J et al (2008) A performance study of general-purpose applications on graphics processors using CUDA. J Parallel Distrib Comput 68(10):1370–1380
Chiosa I, Kolb A (2011) GPU-based multilevel clustering. IEEE Trans Visual Comput Graphics 17(2):132–145
Coates A, Ng AY (2012) Learning feature representations with k-means. In: Neural Networks: Tricks of the Trade. Springer, p 561–580
Drake J, Hamerly G (2012) Accelerated k-means with adaptive distance bounds. In: NIPS Workshop on Optimization for Machine Learning, pp 42–53
Elkan C (2003) Using the triangle inequality to accelerate k-means. In: International Conference on Machine Learning. AAAI Press, pp 147–153
Fahad A, Alshatri N, Tari Z et al (2014) A survey of clustering algorithms for big data: taxonomy and empirical analysis. IEEE Trans Emerg Top Comput 2(3):267–279
Fang W, Lau KK, Lu M, et al (2008) Parallel data mining on graphics processors. Tech. Rep. HKUST-CS08-07, Hong Kong Univ. Sci. and Technology, Hong Kong, China
Farivar R, Rebolledo D, Chan E, et al (2008) A parallel implementation of k-means clustering on GPUs. In: International Conference on Parallel and Distributed Processing Techniques and Applications. CSREA Press, pp 340–345
Hamerly G (2010) Making k-means even faster. In: SIAM International Conference on Data Mining. SIAM, pp 130–140
Hamerly G, Drake J (2015) Accelerating Lloyd’s algorithm for k-means clustering. In: Partitional Clustering Algorithms. Springer, p 41–78
He G, Vialle S, Baboulin M (2022) Parallel and accurate k-means algorithm on CPU-GPU architectures for spectral clustering. Concurr Comput: Pract Exp 34(14):e6621
Hong-Tao B, Li-li H, Dan-tong O, et al (2009) K-means on commodity GPUs with CUDA. In: WRI World Congress on Computer Science and Information Engineering. IEEE Computer Society, pp 651–655
Jain AK (2010) Data clustering: 50 years beyond k-means. Pattern Recogn Lett 31(8):651–666
Jian L, Wang C, Liu Y et al (2013) Parallel data mining techniques on graphics processing unit with compute unified device architecture (CUDA). J Supercomput 64(3):942–967
Kruliš M, Kratochvíl M (2020) Detailed analysis and optimization of CUDA k-means algorithm. In: 49th International Conference on Parallel Processing-ICPP, pp 1–11
Langdon WB (2013) Large-scale bioinformatics data mining with parallel genetic programming on graphics processing units. In: Massively Parallel Evolutionary Computation on GPGPUs. Springer, p 311–347
Lee CC, Chu KY (2012) CUDA-accelerated hierarchical k-means, unpublished manuscript
Li Y, Zhao K, Chu X et al (2013) Speeding up k-means algorithm by GPUs. J Comput Syst Sci 79(2):216–229
Lloyd S (1982) Least squares quantization in PCM. IEEE Trans Inf Theory 28(2):129–137
Lutz C, Breß S, Rabl T et al (2018) Efficient and scalable k-means on GPUs. Datenbank-Spektrum 18(3):157–169
Mittal S, Vetter JS (2015) A survey of CPU-GPU heterogeneous computing techniques. ACM Comput Surv (CSUR) 47(4):69
Mohebi A, Aghabozorgi S, Ying Wah T et al (2016) Iterative big data clustering algorithms: a review. Software: Practice and Experience 46(1):107–129
NVIDIA (2021) CUDA C programming guide. https://docs.nvidia.com/cuda/cuda-c-programming-guide/
Owens JD, Luebke D, Govindaraju NK et al (2005) A survey of general-purpose computation on graphics hardware. In: Conference of the European Association for Computer Graphics. Eurographics Association, pp 21–51
Owens JD, Houston M, Luebke D et al (2008) GPU computing. Proc IEEE 96(5):879–899
Pennington J, Socher R, Manning CD (2021) Global vectors for word representation. https://nlp.stanford.edu/projects/glove/
Phillips SJ (2002) Acceleration of k-means and related clustering algorithms. In: Workshop on Algorithm Engineering and Experiments. Springer, pp 166–177
Sajana T, Rani CS, Narayana K (2016) A survey on clustering techniques for big data mining. Indian J Sci Technol 9(3):1–12
Shirkhorshidi AS, Aghabozorgi S, Wah TY et al (2014) Big data clustering: a review. In: International Conference on Computational Science and Its Applications. Springer, pp 707–720
Taylor C, Gowanlock M (2021) Accelerating the yinyang k-means algorithm using the GPU. In: 2021 IEEE 37th International Conference on Data Engineering (ICDE), IEEE, pp 1835–1840
Upadhyaya SR (2013) Parallel approaches to machine learning - a comprehensive survey. J Parallel Distrib Comput 73(3):284–292
Vassilvitskii S, Arthur D (2006) k-means++: The advantages of careful seeding. In: Annual ACM-SIAM Symposium on Discrete Algorithms. SIAM, pp 1027–1035
Wu R, Zhang B, Hsu M (2009) Clustering billions of data points using GPUs. In: Combined Workshops on Unconventional High Performance Computing Workshop Plus Memory Access Workshop, ACM, pp 1–6
Xu R, Wunsch D (2005) Survey of clustering algorithms. IEEE Trans Neural Netw 16(3):645–678
Yang C, Li Y, Cheng F (2020) Accelerating k-means on GPU with CUDA programming. In: IOP Conference Series: Materials Science and Engineering, IOP Publishing, p 012036
Zechner M, Granitzer M (2009) Accelerating k-means on the graphics processor via CUDA. In: First International Conference on Intensive Applications and Services. IEEE Computer Society, pp 7–15
Funding
Open Access funding enabled and organized by CAUL and its Member Institutions
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors have no relevant financial or non-financial interests to disclose. This research was supported by the University of Waikato.
Additional information
Responsible editor: Johannes Fürnkranz.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Li, M., Frank, E. & Pfahringer, B. Large scale K-means clustering using GPUs. Data Min Knowl Disc 37, 67–109 (2023). https://doi.org/10.1007/s10618-022-00869-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10618-022-00869-6