Abstract
Monitoring the occupancy and abundance of wildlife populations is key to evaluate their conservation status and trends. However, estimating these parameters often involves time and resource-intensive techniques, which are logistically challenging or even unfeasible for rare and elusive species that occur patchily and in small numbers. Hence, surveys based on field identification of signs (e.g. faeces, footprints) have long been considered a cost-effective alternative in wildlife monitoring, provided they produce reliable detectability and meaningful indices of population abundance. We tested the use of sign surveys for monitoring rare and otherwise elusive small mammals, focusing on the Cabrera vole (Microtus cabrerae) in Portugal. We asked how sampling intensity affects true positive detection of the species, and whether sign abundance is related to population size. We surveyed Cabrera voles’ latrines in 20 habitat patches known to be occupied, and estimated ‘true’ population size at each patch using DNA-based capture-recapture techniques. We found that a searching rate of ca. 3 min/250m2 of habitat based on adaptive guided transects was sufficient to provide true positive detection probabilities > 0.85. Sign-based abundance indices were at best moderately correlated with estimates of ‘true’ population size, and even so only for searching rates > 12 min/250m2. Our study suggests that surveys based on field identification of signs should provide a reliable option to estimate occupancy of Cabrera voles, and possibly for other rare or elusive small mammals, but cautions should be exercised when using this approach to infer population size. In case of practical constraints to the use of more accurate methods, a considerable sampling intensity is needed to reliably index Cabrera voles’ abundance from sign surveys.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Monitoring wildlife populations is critical to understand species responses to environmental change, and to inform conservation planning and management (Nichols and Williams 2006; Lindenmayer and Likens 2010). Typically, wildlife monitoring involves estimates of occupancy or population size within an area of interest (e.g. Yoccoz et al. 2001; Mackenzie et al. 2002). Occupancy can be assessed as the proportion of an area containing a species, based on repeated observations of its presence or absence (or more properly detection or non-detection) at several sites (Mackenzie et al. 2002). Higher quality data for estimating population size usually requires more demanding study designs and sophisticated sampling techniques (e.g. Mills et al. 2000; Royle and Young 2008; O’Brien 2011), which is why many studies often use simplified methods providing indices of population abundance (Engeman 2005; Jareño et al. 2014). Methods for estimating occupancy and population size or abundance can provide meaningful information even when a proportion of occupied patches or individuals remains undetected. However, they still pose a number of challenges related to the appropriate monitoring strategies, scales, and sampling techniques considered (e.g. Pollock et al. 2002; Joseph et al. 2006; Steenweg et al. 2018). This is particularly true for rare or elusive species (i.e. patchily distributed species occurring at low abundance, or secretive and difficult-to-observe species, Thompson 2004), for which accurate population monitoring typically requires time and resource intensive techniques (e.g. live-trapping; camera-trapping; non-invasive DNA sampling). These techniques are often difficult and/or costly to implement over large spatial and temporal scales (Witmer 2005; Perkins et al. 2013), and, in the case of rare or hardly detected species, they frequently provide only sparse data, from which robust estimation of demographic parameters often remains challenging (Engeman 2005).
Although surveys based on field identification of signs (e.g., faeces, footprints, hair, dens) cannot deliver information on important population parameters such as age class structure, sex-ratios and reproduction, they are generally considered a cost-effective alternative to more demanding wildlife monitoring techniques for many terrestrial mammals (Wemmer et al. 1996; Wilson and Delahay 2001; Stanley and Royle 2005). However, to provide comparable and informative inferences on population status and trends, sign surveys require the use of explicit and easy-to-replicate field sampling protocols, based on adequate searching strategies and sampling intensity. Low detectability of signs in occupied patches, resulting for instance from inadequate sampling intensity, may lead to bias and imprecision in occupancy estimates (MacKenzie et al. 2002; Ward et al. 2017), and prevent the use of sign surveys for indexing local population abundance (e.g. Rhodes and Jonzén 2011). Therefore, studies assessing the impact of sampling intensity (e.g. duration of surveys and/or spatial coverage of surveyed area), on sign detection success, and on the utility of sign surveys to infer population size are essential for designing efficient monitoring programs, and to correctly interpret ecological and evolutionary processes (e.g. Holbrook et al. 2015; Carreras-Duro et al. 2016; Bowden et al. 2000).
Here we address these issues for the globally near-threatened, Iberian endemic Cabrera vole (Microtus cabrerae), a low-abundance small mammal with fragmented distribution, associated to tall and humid grassy habitat patches, and showing a metapopulation-like spatial structure and dynamics in Mediterranean agricultural landscapes (Pita et al. 2014). Because detectability from life-trapping is relatively low, even under large capture efforts (e.g. Sabino-Marques et al. 2018), Cabrera vole population sampling is often based on sign surveys to infer occupancy and relative abundance within habitat patches (San Miguel 1992; Santos et al. 2006; Pita et al. 2007, 2016; Valerio et al. 2020). However, there are still uncertainties regarding for instance the survey effort required for detecting the species at low densities, as well as on whether sign abundance can be used as a proxy for population size. Notably, it is still largely unknown how sampling intensity may influence species detectability and the strength of inferences regarding population abundance (e.g. Gopalaswamy et al. 2015). Understanding these issues is important for evaluating the effectiveness and potential limitations of surveys based on field identification of signs for monitoring Cabrera vole populations.
We investigated how sampling intensity affects the detection probability (or more precisely the true positive detection probability) of the Cabrera vole in occupied patches from Mediterranean farmland, and whether sign abundance provides a proxy to infer ‘true’ population size at habitat patches. Population size was estimated from capture-recapture (CR) data based on genetic non-invasive sampling (gNIS) of vole faeces, which is known to provide reliable estimates of Cabrera vole’s demographic parameters (Ferreira et al. 2018; Sabino-Marques et al. 2018; Proença-Ferreira et al. 2019). Despite its cost-effectiveness relative to other sampling methods such as live-trapping (see Ferreira et al. 2018), gNIS still requires considerable time and laboratory costs (e.g. DNA extraction kits, PCR, species, sex and individual identification through genotyping faeces) to be easily implemented at large scales (Proença-Ferreira et al. 2019). Testing how much information is lost when using more practical and rapid methods relying solely on field identification of signs, is therefore important for researchers and conservation practitioners (Pita et al. 2014).
Overall, we expect that sampling intensity should have a major overriding effect on true positive detection probability of Cabrera voles based on sign surveys, as well as the strength of relationships between sign abundance indices and population size within habitat patches. Specifically, we expect that detection and inferences on population size should improve with increasing sampling intensity at least up to a certain level beyond which there might be no significant improvements in information quality (e.g. Jones 2011; Reynolds et al. 2011; Green et al. 2020). If true, these expectations will affect the way population monitoring programs based on sign surveys should be implemented in order to maximize information gain and minimize sampling costs (Legg and Nagy 2006; Jones 2011).
Methods
Study area and species
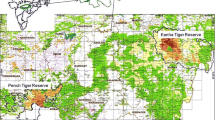
The study was carried out in south-western Portugal (Fig. 1), where Cabrera voles typically occur in small marginal habitat patches (often < 0.2 ha) amid a matrix of unsuitable agricultural habitats, largely dominated by irrigated crops, pastures and greenhouses (Pita et al. 2007). Within patches, voles are typically grouped in subpopulations or colonies consisting of a few individuals (often < 30 animals/ha, Sabino-Marques et al. 2018), which are usually organized as a monogamous breeding pair and their offspring (Pita et al. 2010, 2014). Individuals generally show strong site fidelity, with average home-ranges around 400m2 (Pita et al. 2010). Home-ranges are scent-marked by deposition of faeces in latrines of up to several dozen of faeces, which are thought to be related to individual communication for territory defence and mate advertisement (Gomes et al. 2013).
Study design and sampling
The study focused on 20 habitat patches occupied by Cabrera voles (Fig. 1; Table S1 and Fig. S1, Supplementary Information). Habitat patch sizes ranged between ca. 320 and ca. 3184 m2 and were generally dominated by a dense cover of perennial herbs from the genus Juncus, Carex, Scipus, Agrostis, Festuca, and Briza, among others, together with scattered shrubs mostly from the genus Rubus, Cistus, Ulex, Genista and Ditrichia. Each patch was examined once between December 2016 and February 2018, except during the hottest and driest months (May–August 2017), when population densities and activity of voles tend to be low (Ventura et al. 1998; Pita et al. 2011a; Grácio et al. 2017). In each patch, surveys were resumed within a mean (± SD) 6.0 ± 1.1 days, so as to assume demographic closure of local populations.
Surveys involved initial mapping of habitat boundaries and the characterization of the internal vegetation structure by visually estimating the percentage cover by herbs and shrubs in randomly placed circular plots of 5 m-radius (see e.g. Peralta et al. 2016). Within each plot we also recorded herb and shrub heights in four sampling points orthogonally located at about 2 m from the plot centre. The number of plots considered in each patch was approximately proportional to its size, varying between 2 and 18 (mean = 8.0 ± 4.8), and in each case the mean measurements were taken to represent vegetation structure of the patches.
Because suitable habitat patches in intensively used farmland are easily identified and delimited, these were taken as our fundamental spatial units for sampling sign abundance and estimating population size. Sign surveys were repeated in each patch in three consecutive days in order to obtain a larger sample size and increase the robustness of our findings. Cabrera vole signs are easily identifiable (particularly based on the size, shape and colour of their faeces), and in our study region (as in most of its distribution range) these signs can be hardly confounded with those of other species (e.g. Grarrido-Garcia and Soriguer 2015). In each patch and day, our survey protocol involved intensive searches for Cabrera vole latrines, by slowly walking crouched through the patch and carefully inspecting areas with microhabitats suitable for the species and other more conspicuous signs of vole activity, such as burrows, runways on vegetation, and grass clipping accumulations (see e.g. Santos et al. 2006; Luque-Larena and Lopez 2007; Pita et al. 2011b), where (or near which) latrines tend to be found. This resulted in zigzag-like transects guided by the continuous tracking of those more conspicuous signs, which were then carefully inspected for the presence of latrines. Our sampling protocol is therefore consistent with a guided adaptive sampling (e.g. Ringvall et al. 1998; Maxwell et al. 2012; Ståhl et al. 2000; Pacifici et al. 2016), through which places with conspicuous signs are used as priors to enhance detection effectiveness and get more precise information (in or case, on vole latrines). In these adaptive sign surveys, we considered a latrine as any cluster of ≥ 3 droppings of different ages (indicating reuse at different times), where each dropping is at less than 10 cm from any other dropping from that cluster, and thus at more than 10 cm from any other cluster. We focused on latrines instead of individual droppings because isolated droppings are more rare and do not indicate repeated use of a site by animals (e.g. St-Laurent and Ferron 2008).
Our adaptive sampling protocol was conducted in each patch and day by 2–3 well-trained, experienced observers (DP, TVF, TM) simultaneously searching different parts of the patch, such that virtually the whole habitat surface was thoroughly covered. The total survey duration within a patch (hereafter TSTime, given as sum of searching times by each observer) was similar across days, ranging between 20 and 240 min across patches, largely depending on their size (see Fig. S1 and Table S1 in Supplementary Information). Sign sampling intensity (estimated as the rate between TSTime and patch size) remained, therefore, roughly similar across patches and days, averaging (95% confidence interval) 17.4 (16.4–18.4) min/250m2 (see Tables 1 and S1 in Supplementary Information). We present sampling intensity rates scaled to 250m2 habitat units for ease of understanding and replication across patches of variable size, even though our adaptive sampling protocol does not involve any prior field delimitation of fixed area units within habitat patches.
During sign surveys, genetic non-invasive sampling (gNIS) was performed by collecting vole faeces for species and individual identification (Proença-Ferreira et al. 2019). Specifically, we collected faeces from all latrines that were at least 2 m apart from the nearest collected sample. This strategy was used to increase the chance of recording as many distinct individuals as possible, thereby achieving a reasonable balance between the potential numbers of gNIS-based ‘captures’ and ‘recaptures’ to allow population size estimation (Proença-Ferreira et al. 2019). Each sample consisted of up to 12 of the freshest faeces (mean = 5.11 ± 1.68), collected using sterilized tweezers from each latrine into individual 2 mL microtubes containing 96% alcohol. In most cases (> 90%; see Results), latrines were not completely removed, so they remained potentially detectable by each observer throughout the 3 days survey. Therefore, considering that new faeces were certainly being deposited by voles along the survey days, we assumed no time effects on sign detection probability. During faecal sample collection, chronometers used to record latrine counts were paused and time count restarted when searches were resumed. To increase the probability of detecting individuals and guarantee that enough material was collected for genetic analyses, faecal sample collection in each patch and sampling day often extended beyond the duration of searches directed to latrine counts. In addition, gNIS was further repeated in each patch a few days later (mean of 3.0 + 1.1 days) for collecting additional faecal samples for genetic identification. While we used roughly the same reference sampling intensity in these additional surveys, the searching pattern strategy was not consistent with the guided sampling protocol used in previous days, often involving additional inspections at lower quality microhabitats (e.g. drier and less vegetated spots). Therefore, we did not use these additional gNIS surveys to derive latrine counts. All faecal samples were stored at − 20 °C until DNA extraction and individual genotyping. The number of captures and recaptures of individual genotypes in each patch were then used to estimate local population size (e.g. Sabino-Marques et al. 2018; Proença-Ferreira et al. 2019), and to assess how these relate with different sign abundance indices.
Sign abundance indices
Sign abundance indices in each habitat patch were directly obtained from latrine counts, a method frequently used to estimate the relative abundance of small mammals, including voles (e.g. Woodroffe et al. 1990; Bonesi et al. 2002). In addition, in order to integrate information on the spatial distribution of latrines (e.g. Lambin et al. 2000), besides latrine counting, we also estimated the extent of the area where vole latrines were found within patches (hereafter, extent of occurrence), as described in Pita et al. (2016). Briefly, the procedure consists in creating and merging 10 m radius buffers centred on each latrine location (see also Poccock et al. 2003), so defined to provide circular areas close to the mean home range size of the species in the study area (Pita et al. 2010).
To assess the influence of sampling intensity on the suitability of sign surveys for inferring local population size, we calculated the number of latrines that would be detected in each patch and day under shorter survey time durations (e.g. Green et al. 2020). Specifically, for each patch and day, we resampled the original survey data after reducing TSTime to 90%, 80%, 70%, 60%, 50%, 40%, 30%, 20% and 10%. This resulted in a total 10 measurements of latrine counts and extent of occurrence per patch and day, overall comprising a tenfold change in sampling intensity (see Tables 1, and S1 in Supplementary Information).
DNA extraction and genotyping
DNA was extracted from faecal samples using the E.Z.N.A.® Tissue DNA Kit (OMEGA bio-tek) following the manufactures instructions, with an initial digestion step using a lysis washing buffer (Maudet et al. 2004) for 15 min at 56 °C. Only faecal samples with potential for being successfully genotyped, as judged by their apparent freshness, were considered for analysis, with a maximum number of ca. 100 samples per patch. Selected samples were genotyped for a set of 11 microsatellites following a stepwise approach, which involved an initial screening for sample quality based on a set of three loci (see Ferreira et al. 2018 for genotyping details). Samples that failed to amplify this first set were discarded from subsequent analyses. Samples that amplified well were then tested for the remaining set of 8 microsatellites (see Table S2, Supplementary Information). Previous results showed that the set of 11 microsatellites are highly informative in providing accurate individual identification and diversity values (Ferreira et al. 2018). Species ID was confirmed using a small fragment of mitochondrial DNA, Dloop (Alasaad et al. 2011). The samples were also sexed using two small-sized sex chromosome introns (DBX5-S and DBY7-S, Ferreira et al. 2018). To account for genotyping errors (e.g. allele dropout and false alleles) and to obtain a consensus genotype, each multiplex reaction was replicated four times (three times for the sex chromosome introns amplification). PCR reactions were performed in a final volume of 10 μL, consisting of 4 μL of Qiagen© Multiplex PCR Kit Master Mix, 1μL of DNA, and primer concentrations and thermal profiles according to Ferreira et al. (2018). All products were sequenced on an ABI3130 Capillary Sequencer (Applied Biosystems). The extractions and PCR reactions of the non-invasive samples were performed in physically isolated rooms, and all the equipment used was sterilized with bleach and ethanol and exposed to UV light before and after usage. Aerosol-resistant pipette tips were used, and negative controls were included in each manipulation, maintaining conditions to monitor and reduce risk of DNA contamination (Beja-Pereira et al. 2009; Barbosa et al. 2013; Costa et al. 2017).
Allele calling of the microsatellite loci and sex chromosome introns were performed using GeneMapper (v.4.0; Applied Biosystems), while Dloop sequences were analysed with Geneious (v.8.0; Kearse et al. 2012). Consensus genotypes for each sample were obtained by analysing all replicate genotypes with the software Gimlet (v.1.3.3, Valière 2002). For genotypes differing by up to two loci or with up to two missing data, additional PCR replicas were performed, to try to complete the genotypes, and check for genotyping errors. Genotyping error rates were estimated using the software Pedant (Johnson and Haydon 2007), with 10,000 search steps. Since the software only allows the comparison of two replicates, all possible pairwise comparisons were performed and the results were averaged. Sample consensus genotypes were then compared with each other to identify individuals. The criteria used to assign samples to individuals was very strict, with only individuals differing in more than two alleles assigned as new captures. Less strict criteria were not evaluated here, as these have little impact in population size estimates of the species (Sabino-Marques et al. 2018). The expected heterozygosity (HE) and observed heterozygosity (HO) for each locus were calculated using the software GenAlEx, and overall inbreeding (FIS) was estimated in the program INEST 2.0 (Chybicki and Burczyk 2009) using 2 × 105 iterations, with 50 iterations of thinning and a burn-in of 2 × 104 iterations (Ferreira et al. 2018).
Population size estimates
To assess Cabrera vole population size at each habitat patch, we pooled the gNIS-based estimated number of genotypes from each sampling day into a single session. We then used the Eggert accumulation curve for capture-recapture (CR) data (Eggert et al. 2003), which is based on the exponential function given as \(\left.E\left(x\right)=a(1-{e}^{\left(bx\right)}\right)\), where \(x\) represents the number of genotyped samples, \(E\left(x\right)\) is the cumulative number of unique genotypes found in \(x\) genotyped samples, \(a\) is the asymptote of the function that represents the estimated population size, and \(b\) is the non-linear slope of the function. The Eggert estimator performs relatively well compared to other popular accumulation curves such as the hyperbolic curve proposed by Kohn et al. (1999), which tends to overestimate population size and result in less precise estimates (Eggert et al. 2003; Frantz et al. 2004), as confirmed in preliminary analyses of our data (not shown here). We used this approach rather than closed-CR because the small sample sizes from most habitat patches prevented the use of algorithms directly estimating recapture probability (e.g. Lukacs and Burnham 2005). Since the order in which the identified genotypes are added may influence the shape of the accumulation curves (Eggert et al. 2003), we randomized each dataset 100 times, and fit the equations to Eggert’s curve using least squares regressions. Estimates of \(a\) (population size) for each dataset were taken as the average of all replicates, and respective point estimates were used as reference of the ‘true’ population size in each patch.
Modelling
We used Generalised Linear Mixed Effects Models (GLMM) implemented in the ‘lme4’ R package (Bates et al. 2015; R Core Team 2020) to model species detection probability based on detection/non-detection data of the species across different sampling intensities, and to assess the effects of sampling intensity on the relationships between population size estimates and abundance indices derived from latrine counts.
We considered detection probability as the probability that a species will be detected at a site, given its presence, i.e. the probability that a species both occupies and is detected in a survey (as in Mackenzie et al. 2002, Mackenzie and Royle 2005), which essentially corresponds to true positive detection probability. This definition assumes that species are never falsely detected (no false positives) and that they may or may be not detected at a site when present (true positive and false negatives, respectively) (Mackenzie et al. 2002). Given that false negative detection probability is the complement of the true positive detection probability, and that false positive detection probability is the complement of true negative detection probability (e.g. Miller et al. 2013), when assuming no false positives in a species survey, the true positive detection may fairly describe detection uncertainty (Mackenzie et al. 2002). In our study, because the data was conditioned to occupied patches, and a non-detection was assumed to represent the overlooking of vole signs, rather than a true absence, we directly modelled detection events using GLMM, and did not need to account for possible non-occurrence based on a site-occupancy model (MacKenzie et al. 2002; see e.g. Chen et al. 2009).
True-positive detection probability of vole latrines was modelled using binomial error distribution (logit link function) and considering the maximal random structure effects justified by our sampling design, so as to better control variation, increase the power of the analyses, and optimize generalization of the findings (e.g. Gillies et al. 2006). Therefore, we included in the random component the patch and the month of sampling, as well as the identity of the observer that first detected Cabrera signs in each patch and day. We then built a set of models including as fixed factors the main and additive effects of sampling intensity and the variables describing vegetation structure within patches, which may also affect true detection probability (e.g. higher shrub cover may prevent or retard the progression of observers across the habitat and therefore affect sign searching efficiency), while also considering the model including only the random effects (null model). To avoid multicollinearity among vegetation variables, we used a principal component analysis (PCA) to quantify main patterns of variation among patches, and used the results as predictors of true-positive detections. We implemented the PCA using the ‘prcomp’ function in R and used the Kaiser criterion (Legendre and Legendre 2012) to keep only principal component axes with associated eigenvalues > 1. We then performed a varimax rotation of the significant PCA axes using principal function in the R package ‘psych’ (Revelle 2015).
The support of each candidate model was based on the Akaike Information Criteria corrected for small samples (AICc; Burnham and Anderson 2002), with ΔAICc < 2 indicating equally supported models (Burnham and Anderson 2002). We also assessed the conditional probability of each model being the best model by estimating the respective AICc-weighs (Burnham and Anderson 2002). The goodness-of-fit of the best model was assessed by computing marginal and conditional R2 for GLMM (Nakagawa and Schielzeth 2013), using the MuMIn package (Barton 2018). Marginal R2 shows the proportion of variance explained by the fixed effects, while conditional R2 provides the proportion of variance explained by both fixed and random effects.
The relationship between population size estimates (rounded to the nearest integer and taken as the dependent variable) and each sign abundance index calculated for each sampling day under variable sampling intensities (fixed effects) was modelled with the negative binomial distribution (log link function), as observations were overdispersed with respect to the Poisson distribution (null models residual deviance decreased from 334.7 to 306.8). Initial models included sampling month and patch as random effects. However, patch effects were removed from the random structure to avoid non-singular fit (Bates et al. 2015). Models including each abundance index were compared with the respective null model based on AICc (ΔAICc and AICc-weighs). Model fit was assessed by computing marginal and conditional R2 for GLMM.
Results
We counted a total of 1409 latrines in the 20 habitat patches surveyed across the 3 consecutive sampling days, with a mean of 23.5 ± 10.8 latrines per patch per day (range: 2–54), and a mean extent of occurrence of 1388.73 ± 645.15 m2 per patch per day (318–2856 m2). A total of 1914 faecal samples were collected for downstream genetic analysis, with 95.7 ± 38.43 (54–217) samples per patch. A total of 1553 samples (77.65 ± 18.85 per patch [47–107]) were analysed, of which 468 samples (23.4 ± 12.17 per patch [9–46]) were successfully genotyped, thus resulting in a mean genotyping success of 29.65% ± 11% (9–46%) (see Table S3, Supplementary Information). The genotyping error rates were low, with an average dropout rate of 1.8 × 10−2 ± 9.9 × 10−3 (3.3 × 10−3–3.1 × 10−2) and a false allele rate of 5.0 × 10−4 ± 8.0 × 10−4 (0.0–2.1 × 10−3) (see Table S4, Supplementary Information).
We genotyped a total of 101 different Cabrera voles (58 females and 43 males), with 5.01 ± 2.96 (1–12) individuals genotyped per patch. Population size estimates based on accumulation curves were computed for all but one patch, which appeared to be occupied only by a solitary male that was genotyped 46 times (see Table S3, Supplementary Information). Overall, the population size estimates obtained through Eggert’s accumulation curve were notably close to the number of vole genotypes enumerated through gNIS, totalling 118.3 ± 4.0 animals, with 6.0 ± 4.1 (1–16) individuals per patch (see Table S3 and Fig. S2, Supplementary Information).
The PCA on variables describing vegetation structure produced one single principal component with eigenvalue of 2.13 and explaining 53% of the variation in vegetation data. This principal component (PC-Veg) described a gradient of vegetation structure contrasting patches with higher shrub cover and high with those largely dominated by a well-developed herbaceous layer (Table 2), and was considered, together with sampling intensity, as predictor of true positive detection probability. From the set of four candidate models (Table 3), the one including the effect of sampling intensity received greatest support, with an AICc more than 4 units lower than the second most supported, which also included PC-Veg (Table 3). The top ranked model returned a marginal R2 of 94%, and revealed significant positive effects of sampling intensity in true positive detection probability (Table 3). This model suggested that a sampling intensity rate of ca. 3 min/250m2 provides a true positive detection probability always > 0.85 (L95%CI), while shorter searching times per unit area result in lower and much more variable detectability of the species when present. According to this model, sampling intensities higher than ca. 6 min/250m2 can virtually achieve perfect detectability of species true presence in a given patch (Fig. 2), suggesting that non detections under such sampling intensity should represent true negatives.
Sign abundance indices and estimates of local population size were in general positively correlated, although the strength of correlation was strongly dependent on sampling intensity. In particular, our results suggest that correlation between abundance indices and population size gets stronger with increasing sampling intensity up to searching rates around 12 min/250m2, tending to stabilize or increase much slowly thereafter (Table 4). Both indices provided moderately strong relationships with estimates of population size only under greater sampling intensity, with slopes reaching 0.34 ± 0.06 and 0.40 ± 0.06 in the case of latrine counts and extent of occurrence, respectively (Table 3). In addition, compared to latrine counts, the extent of occurrence explained a higher proportion of the variance in population size estimates, with marginal R2 reaching 0.31 and 0.39, respectively (Table 4, Figs. 3, and S3, Supplementary Information).
Predicted population size of Cabrera voles according to latrine counts (a) and extent of occurrence (b) resulting from the highest sampling intensity (SI-10, mean [95%CI] = 17.4 [16.4–18.4] min/250 m2, see Table 1). In each case, lines are mean values; grey areas are 95% confidence intervals; circles are observed values (see also Fig. S3 in Supplementary Information)
Discussion
The development of cost-effective methods based on field identification of animal signs for monitoring wildlife population has been for long a priority in ecological and conservation studies (Witmer 2005). However, for most species such methods are largely lacking or, when available, they are often poorly calibrated and tested (e.g. Hopkins and Kennedy 2004; Gervais 2010), making it difficult to properly infer population changes, determine species status, and inform conservation management (Thompson 2004). Focusing on Cabrera voles, our study provides evidence that, where other vole species producing similar signs are absent (i.e. no false positives are likely to occur), and when other more accurate methods like live-trapping or gNIS are not available, sign surveys may provide useful low-cost alternative for monitoring occupancy and inferring local population abundance. However, our study also showed that, when employing sampling protocols based on continuous zigzag-like tracking paths adapted to improve vole latrine detectability, the rate of time spent searching for signs per unit area, strongly affects the reliability of this method. This suggests that differences in population size indices may arise due to differences in sampling intensity, so affecting the quality of inferences on population status and trends (Holbrook et al. 2015; Carreras-Duro et al. 2016; Bowden et al. 2000). Our study thus supports the idea that standardized survey protocols and time-based sampling intensities designed to enhance species detectability and population size indexing from sign surveys are needed to provide comparable and informative population assessments of Cabrera voles, and other similar species, across space and time (Pollock et al. 2002; Yoccoz et al. 2001). Therefore, we believe our approach provides important insights on how sign-based population monitoring should be implemented in a cost-effective way, particularly as regards to optimal allocation of survey time duration, which is critical to design and planning monitoring studies over large spatial and temporal scales.
Cabrera vole detectability
Cabrera vole population assessments are often limited to single-visit presence-absence surveys within suitable habitat patches, based on sign searches conducted within short (though often poorly defined) time intervals (e.g. Santos et al. 2006; Pita et al. 2007; Valerio et al. 2020). Although such surveys may raise concerns regarding detectability issues, our study confirmed that where the species is present, its signs may be readily detected within the very first sampling minutes, suggesting that this method provides a reliable approach for studying species distribution and occupancy patterns, at least where other species producing similar signs are absent (no false positives). However, our results also showed that, according to our expectations, sampling intensity had a major influence on true positive detection probability of Cabrera vole signs, overriding other eventual sources of variability, such as vegetation structure. This suggests that careful consideration is needed regarding the sampling intensity employed, in order to enhance sign detection probability, and the use of sign-surveys in monitoring programs either based on species detection histories. While standard methods accounting for observation error in occupancy modelling can efficiently deal with imperfect detection, low true positive detection rates may result in less accurate and precise estimates of species occupancy (MacKenzie et al. 2002; Mackenzie and Royle 2005), thus making the choice of the sampling intensity a critical step in such studies. Several studies have recommended that the survey duration in occupancy studies should be considered in a way that the probability of detection exceeds 0.8. (e.g. Long et al. 2008; Steenveg et al. 2018). Our results indicated that an experienced observer surveying at a sampling intensity rate of up to 3 min/250m2 of suitable habitat will likely achieve a true positive detectability > 0.85 (L95%CI), suggesting that this sampling intensity could be used as a reference threshold for guiding vole sign surveys based on detection-non detection data, assuming no false positives. Sampling intensities up to 6 min/250m2 apparently guarantee a virtually perfect true positive detectability. We stress, however, that achieving nearly perfect detectability for monitoring occupancy may be desirable only in studies relying on naïve occupancy estimates, or single location surveys focused for instance on determining whether voles are present at a given patch where some management activity is likely to affect local habitat quantity or quality (de Solla et al. 2005). Otherwise, for distributional studies conducted over large scales, a lower level of detectability involving sampling intensities of up to 3 min/250m2, may be fairly tolerated due to the trade-offs between maximizing the number of total locations that can be surveyed versus the time spent surveying at each location (MacKenzie and Royle 2005).
Population size indices
Our results suggest that both latrine counts and the estimated area used by voles within patches (extent of occurrence) correlated positively with population size estimates. The strength of such relationships was weak for low sampling intensities, but increased along with survey durations of up to ca. 12 min/250m2, tending to stabilize at moderate levels thereafter. This indicates that sign surveys based on guided sampling under low sampling intensity should not be used for indexing Cabrera vole population size. The optimal sampling intensity should thus be around ca. 12 min/250m2, above which there may be limited gains in population size information. Our results also indicate that the spatial extent of latrines (extent of occurrence) within patches (Pita et al. 2016) is more strongly related than latrine counts to the estimates of population size, and so the former might be preferred in Cabrera vole monitoring. This result is probably because the estimated area occupied by vole integrates information on the spatial distribution of latrines (e.g. Lambin et al. 2000), thus likely accounting to some extent for sources of variability related to non-uniform distribution of voles within patches (St-Laurent and Ferron 2008), individual variations in marking behaviour within occupied territories (e.g. Ferkin et al. 2004), or possible individual heterogeneity in sign detection between individuals (Watkins et al. 2010).
Despite the observation of a significant relation of Cabrera vole abundance with both latrine counts and extent of occurrence, the magnitude of such relation was at best moderate. Reasons for this are uncertain, but besides the individual variations and social context that may affect individuals’ spatial marking behaviours (Ferkin et al. 2004), the uncertainties in population size estimates based on gNIS and asymptotic estimators may have also affected the strength of relationships found. Although gNIS was based on an optimized protocol for reducing genotyping error rates (Ferreira et al. 2018), a large number of samples that were extracted did not produce results for one or more microsatellites, or were contaminated, resulting in a relatively low genotyping success which can affect population estimates (Waits and Leberg 2000; Luikart et al. 2010). On the other hand, asymptotic approaches provide the simplest population size estimators from CR data, relying on overly naive assumptions (e.g. random spatial distribution, homogeneous capture probabilities in space and time) that are seldom found in natural systems (Waits and Leberg 2000; Miller et al. 2005; Luikart et al. 2010). However, since the total number of individuals identified in each patch was very close to the estimates produced by asymptotic estimators, we assumed that our sampling procedures and rarefaction-based methods provided a reliable basis to infer local population size, potentially allowing the identification of most (if not all) the individuals present in each patch. Nearly complete censuses are considered a useful approach for inferring abundance of small populations (Gerber et al. 2014), because traditional CR-based estimation methods are difficult to apply at the patch level, as previously shown for the Cabrera vole (e.g. Fernández-Salvador et al. 2005; Ferreira et al. 2018; Sabino-Marques et al. 2018).
Conclusions and implications
Overall, our study suggests that whenever field sign identification is made with negligible error (no false positives), and other more accurate tools are not available, sign searches by experienced observers may provide an adequate alternative to help understanding the distribution changes and occupancy dynamics of small mammals like the Cabrera vole, making this method suitable for population monitoring across large spatial and temporal scales (e.g. Pita et al. 2007, 2016). Furthermore, our work also shows that despite being potentially time-consuming, sign surveys based on the abundance indices analysed in this study may also provide a useful approximation to infer Cabrera vole local population size, at least in systems where habitat patches are mostly small, easily recognised and delimited, and where other vole species producing similar signs are absent, as it is the case of our study area. However, whenever detailed information on local population size is needed, gNIS combined with CR modelling should provide a more adequate approach (e.g. Sabino-Marques et al. 2018; Proença-Ferreira et al. 2019). We thus stress that the decision of whether to use or not abundance indices in any particular study based on sign searches should involve a cost–benefit analysis accounting for the specific study objectives (e.g. Pollock et al. 2002; Falcy et al. 2016; Ferreira et al. 2018).
Data Availability
Data are available from the corresponding author upon reasonable request.
References
Alasaad S, Soriguer RC, Jowers MJ, Marchal JA, Romero I, Sánchez A (2011) Applicability of mitochondrial DNA for the identification of Arvicolid species from faecal samples: A case study from the threatened Cabrera’s vole. Mol Ecol Res 11:409–414. https://doi.org/10.1111/j.1755-0998.2010.02939.x
Barbosa S, Pauperio J, Searle JB, Alves PC (2013) Genetic identification of Iberian rodent species using both mitochondrial and nuclear loci: Application to noninvasive sampling. Mol Ecol Res 13:43–56. https://doi.org/10.1111/1755-0998.12024
Barton K (2018) Package ‘MuMIn’. R package version 1.46. R Foundation for Statistical Computing, Vienna. https://cran.r-project.org/web/packages/MuMIn/index.html. Accessed 22 May 2022
Bates D, Mächler M, Bolker BM, Walker SC (2015) Fitting linear mixed-effects models using lme4. J Stat Softw 67:1–48. https://doi.org/10.18637/jss.v067.i01
Beja-Pereira A, Oliveira R, Alves PC, Schwartz MK, Luikart G (2009) Advancing ecological understandings through technological transformations in noninvasive genetics. Mol Ecol Res 9:1279–1301. https://doi.org/10.1111/j.1755-0998.2009.02699.x
Bonesi L, Rushton S, Macdonald D (2002) The combined effect of environmental factors and neighbouring populations on the distribution and abundance of Arvicola terrestris. An approach using rule-based models. Oikos 99(2):220–230. https://doi.org/10.1034/j.1600-0706.2002.990202.x
Bowden D, White GC, Bartmann RM (2000) Optimal allocation of sampling effort for monitoring a harvested mule deer population. J Wildl Manage 64:1013–1024. https://doi.org/10.2307/3803212
Burnham KP, Anderson DR (2002) Model selection and inference: A practical information-theoretic approach, 2nd edn. Springer-Verlag, New York
Carreras-Duro J, Moleón M, Barea-Azcón JM, Ballesteros-Duperón E, Virgós E (2016) Optimization of sampling effort in carnivore surveys based on signs: a regional scale study in a Mediterranean area. Mamm Biol 81:205–213. https://doi.org/10.1016/j.mambio.2015.12.003
Chen GK, Kéry M, Zhang JL, Ma KP (2009) Factors affecting detection probability in plant distribution studies. J Ecol 97:1383–1389. https://doi.org/10.1111/j.1365-2745.2009.01560.x
Chybicki IJ, Burczyk J (2009) Simultaneous estimation of null alleles and inbreeding coefficients. J Hered 100:106–113. https://doi.org/10.1093/jhered/esn088
Costa V, Rosenbom S, Monteiro R, O’Rourke SM, Beja-Pereira A (2017) Improving DNA quality extracted from fecal samples - a method to improve DNA yield. Eur J Wildl Res 63:3. https://doi.org/10.1007/s10344-016-1058-1
de Solla SR, Shirose LJ, Fernie KJ, Barrett GC, Brousseau CS, Bishop CA (2005) Effect of sampling effort and species detectability on volunteer based anuran monitoring programs. Biol Conserv 121:585–594. https://doi.org/10.1016/j.biocon.2004.06.018
Eggert LS, Eggert JA, Woodruff DS (2003) Estimating population sizes for elusive animals: The forest elephants of Kakum National Park, Ghana. Mol Ecol 12:1389–1402. https://doi.org/10.1046/j.1365-294X.2003.01822.x
Engeman RM (2005) Indexing principles and a widely applicable paradigm for indexing animal populations. Wildl Res 32:203–210. https://doi.org/10.1071/WR03120
Falcy MR, McCormick JL, Miller SA (2016) Proxies in practice: calibration and validation of multiple indices of animal abundance. J Fish Wildl Manag 7:117–128. https://doi.org/10.3996/092015-JFWM-090
Ferkin MH, Lee DN, Leonard ST (2004) The reproductive state of female voles affects their scent marking behavior and the responses of male conspecifics to such marks. Ethology 110:257–272. https://doi.org/10.1111/j.1439-0310.2004.00961.x
Fernández-Salvador R, Ventura J, García-Perea R (2005) Breeding patterns and demography of a population of the Cabrera vole. Microtus Cabrerae Anim Biol 55:147–161. https://doi.org/10.1163/1570756053993497
Ferreira CM, Sabino-Marques H, Barbosa S, Costa P, Encarnação C, Alpizar-Jara R, Pita R, Beja P, Mira A, Searle JB, Paupério J, Alves PC (2018) Genetic non-invasive sampling (gNIS) as a cost-effective tool for monitoring elusive small mammals. Eur J Wildl Res 64:46. https://doi.org/10.1007/s10344-018-1188-8
Frantz AC, Schaul M, Pope LC, Fack F, Schley L, Muller CP, Roper TJ (2004) Estimating population size by genotyping remotely plucked hair: The Eurasian badger. J Appl Ecol 41:985–995. https://doi.org/10.1111/j.0021-8901.2004.00951.x
Garrido-García JA, Soriguer RC (2015) Topillo de Cabrera Iberomys cabrerae (Thomas, 1906). Guía de indícios de los mamíferos de España. SECEM. http://www.secem.es/wp-content/uploads/2015/07/020-Iberomys-cabrerae.pdf
Gerber BD, Ivan JS, Burnham KP (2014) Estimating the abundance of rare and elusive carnivores from photographic-sampling data when the population size is very small. Popul Ecol 56:463–470. https://doi.org/10.1007/s10144-014-0431-8
Gervais JA (2010) Testing sign indices to monitor voles in grasslands and agriculture. Northwest Sci 84:282–288. https://doi.org/10.3955/046.084.0307
Gillies CS, Hebblewhite M, Nielsen SE, Krawchuk MA, Aldridge CL, Frair JL, Saher DJ, Stevens CE, Jerde CL (2006) Application of random effects to the study of resource selection by animals. J Anim Ecol 75:887–898. https://doi.org/10.1111/j.1365-2656.2006.01106.x
Gomes LAP, Mira APP, Barata EN (2013) The role of scent-marking in patchy and highly fragmented populations of the Cabrera Vole (Microtus cabrerae Thomas. Zool Sci 30(4):248–254. https://doi.org/10.2108/zsj.30.248
Gopalaswamy A, Delampady M, Karanth KU, Kumar NS, Macdonald DW (2015) An examination of index-calibration experiments: counting tigers at macroecological scales. Methods Ecol Evol 6(9):1055–1066. https://doi.org/10.1111/2041-210X.12351
Grácio AR, Mira A, Beja P, Pita R (2017) Diel variation in movement patterns and habitat use by the Iberian endemic Cabrera vole: implications for conservation and monitoring. Mamm Biol 83:21–26. https://doi.org/10.1016/j.mambio.2016.11.008
Green NS, Wildhaber ML, Albers JL, Pettit TH, Hooper MJ (2020) Efficient mammal biodiversity surveys for ecological restoration monitoring. Integr Environ Assess Manag. https://doi.org/10.1002/ieam.4324
Holbrook JD, Arkle RS, Rachlow JL, Vierling KT, Pilliod DS (2015) Sampling animal sign in heterogeneous environments: how much is enough? J Arid Environ 199:51–55. https://doi.org/10.1016/j.jaridenv.2015.03.013
Hopkins HL, Kennedy ML (2004) An assessment of indices of relative and absolute abundance for monitoring populations of small mammals. Wildl Soc Bull 32:1289–1296. https://doi.org/10.2193/0091-7648(2004)032[1289:AAOIOR]2.0.CO;2
Jareño D, Viñuela J, Luque-Larena JJ, Arroyo L, Arroyo B, Mougeot F (2014) A comparison of methods for estimating common vole (Microtus arvalis) abundance in agricultural habitats. Ecol Indic 36:111–119. https://doi.org/10.1016/j.ecolind.2013.07.019
Johnson PCD, Haydon DT (2007) Maximum-likelihood estimation of allelic dropout and false allele error rates from microsatellite genotypes in the absence of reference data. Genetics 175:827–842. https://doi.org/10.1534/genetics.106.064618
Jones JPG (2011) Monitoring species abundance and distribution at the landscape scale. J Appl Ecol 48:9–13. https://doi.org/10.1111/j.1365-2664.2010.01917.x
Joseph LN, Field SA, Wilcox C, Possingham HP (2006) Presence–absence versus abundance data for monitoring threatened species. Conserv Biol 20:1679–1687. https://doi.org/10.1111/j.1523-1739.2006.00529.x
Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, Thierer T, Ashton B, Meintjes P, Drummond A (2012) Geneious Basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28:1647–1649. https://doi.org/10.1093/bioinformatics/bts199
Kohn MH, York EC, Kamradt DA, Haught G, Sauvajot RM, Wayne RK (1999) Estimating population size by genotyping faeces. Proc Biological Sci B 266:657–663. https://doi.org/10.1098/rspb.1999.0686
Lambin X, Petty SJ, MacKinnon JL (2000) Cyclic dynamics in field vole populations and generalist predation. J Anim Ecol 69:106–118. https://doi.org/10.1046/j.1365-2656.2000.00380.x
Legendre P, Legendre L (2012) Numerical Ecology. Third English edition. Elsevier Science BV, Amsterdam, The Netherlands
Legg CJ, Nagy L (2006) Why most conservation monitoring is, but need not be, a waste of time. J Environ Manage 78:194–199. https://doi.org/10.1016/j.jenvman.2005.04.016
Lindenmayer DB, Likens GE (2010) The science and application of ecological monitoring. Biol Conserv 143:1317–1328. https://doi.org/10.1016/j.biocon.2010.02.013
Long R, Zielinski WJ, Long R, MacKay P, Zielinski W, Ray J (2008) Designing effective noninvasive carnivore surveys. In: Long RA, MacKay P, Ray JC, Zielinski WJ (eds) Noninvasive survey methods for carnivores. Island Press, Washington, D.C., USA, pp 8–44
Luikart G, Ryman N, Tallmon DA, Schwartz MK, Allendorf FW (2010) Estimation of census and effective population sizes: The increasing usefulness of DNA-based approaches. Conserv Genet 11:355–373. https://doi.org/10.1007/s10592-010-0050-7
Lukacs PM, Burnham KP (2005) Estimating population size from DNA‐based closed capture‐recapture data incorporating genotyping error. J Wildl Manage, 69:396–403. https://doi.org/10.2193/0022-541X(2005)069<0396:EPSFDC>2.0.CO;2
Luque-Larena JJ, López P (2007) Microhabitat use by wild-ranging Cabrera voles Microtus cabrerae as revealed by live trapping. Eur J Wildl Res 53:221–225. https://doi.org/10.1007/s10344-006-0084-9
MacKenzie DI, Nichols JD, Lachman GB, Droege S, Royle JA, Langtimm CA (2002) Estimating site occupancy rates when detection probabilities are less than one. Ecology 83:2248–2255. https://doi.org/10.1890/0012-9658(2002)083[2248:ESORWD]2.0.CO;2
Mackenzie DI, Royle JA (2005) Designing occupancy studies: General advice and allocating survey effort. J Appl Ecol 42:1105–1114. https://doi.org/10.1111/j.1365-2664.2005.01098.x
Maudet C, Luikart G, Dubray D, Von Hardenberg A, Taberlet P (2004) Low genotyping error rates in wild ungulate faeces sampled in winter. Mol Ecol Notes 4(4):772–775. https://doi.org/10.1111/j.1471-8286.2004.00787.x
Maxwell BD, Backus V, Hohmann MG, Irvine KM, Lawrence P, Lehnhoff EA, Rew LJ (2012) Comparison of transect-based standard and adaptive sampling methods for invasive plant species. Invasive Plant Sci Manag 5:178–193. https://doi.org/10.1614/IPSM-D-11-00022.1
Miller CR, Joyce P, Waits LP (2005) A new method for estimating the size of small populations from genetic mark-recapture data. Mol Ecol 14:1991–2005. https://doi.org/10.1111/j.1365-294X.2005.02577.x
Miller DAW, Nichols JD, Gude JÁ, Rich LN, Podruzny KM, Hines JE, Mitchell MS (2013) Determining occurrence dynamics when false positives occur: Estimating the range dynamics of wolves from public survey data. PLoS ONE 8:e65808. https://doi.org/10.1371/journal.pone.0065808
Mills L, Citta J, Lair K, Schwartz M, Tallmon D (2000) Estimating animal abundance using non-invasive DNA sampling: promise and pitfalls. Ecol Appl 10:283–294. https://doi.org/10.1890/1051-0761(2000)010[0283:EAAUND]2.0.CO;2
Nakagawa S, Schielzeth H (2013) A general and simple method for obtaining R2 from generalized linear mixed-effects models. Methods Ecol Evol 4:133–142. https://doi.org/10.1111/j.2041-210x.2012.00261.x
Nichols JD, Williams BK (2006) Monitoring for conservation. Trends Ecol Evol 21:668–673. https://doi.org/10.1016/j.tree.2006.08.007
O’Brien TG (2011) Abundance, density and relative abundance: a conceptual framework. In: O’Connell AF, Nichols JD, Karth KU (eds) Camera Traps in Animal Ecology. Springer, New York city, New York, USA, pp 71–96
Pacifici K, Reich BJ, Dorazio RM, Conroy MJ (2016) Occupancy estimation for rare species using a spatially-adaptive sampling design. Methods Ecol Evol 7:285–293. https://doi.org/10.1111/2041-210X.12499
Peralta D, Leitão I, Ferreira A, Mira A, Beja P, Pita R (2016) Factors affecting southern water vole (Arvicola sapidus) detection and occupancy probabilities in Mediterranean farmland. Mamm Biol 82(2):123–129. https://doi.org/10.1016/j.mambio.2015.10.006
Perkins GC, Kutt AS, Vanderduys EP, Perry JJ (2013) Evaluating the costs and sampling adequacy of a vertebrate monitoring program. Aust Zool 36:373–380. https://doi.org/10.7882/AZ.2013.003
Pita R, Beja P, Mira A (2007) Spatial population structure of the Cabrera vole in Mediterranean farmland: The relative role of patch and matrix effects. Biol Conserv 134(3):383–392. https://doi.org/10.1016/j.biocon.2006.08.026
Pita R, Lambin X, Mira A, Beja P (2016) Hierarchical spatial segregation of two Mediterranean vole species: the role of patch-network structure and matrix composition. Oecologia 182(1):253–263. https://doi.org/10.1007/s00442-016-3653-y
Pita R, Mira A, Beja P (2010) Spatial segregation of two vole species (Microtus cabrerae and Arvicola sapidus) within habitat patches in a highly fragmented farmland landscape. Eur J Wildl Res 56:651–556. https://doi.org/10.1007/s10344-009-0360-6
Pita R, Mira A, Beja P (2011a) Circadian activity rhythms in relation to season, sex and interspecific interactions in two Mediterranean voles. Anim Behav 81:1023–1030. https://doi.org/10.1016/j.anbehav.2011.02.007
Pita R, Mira A, Beja P (2011b) Assessing habitat differentiation between coexisting species: The role of spatial scale. Acta Oecol 37:124–132. https://doi.org/10.1016/j.actao.2011.01.006
Pita R, Mira A, Beja P (2014) Microtus cabrerae (Rodentia: Cricetidae). Mamm Species 912(912):48–70. https://doi.org/10.1644/912.1
Poccock MJO, White PCL, McClean CJ, Searle JB (2003) The use of accessibility in defining sub-groups of small mammals from point sampled data. Comput Environ Urban 27:71–83. https://doi.org/10.1016/S0198-9715(01)00037-0
Pollock KH, Nichols JD, Simons TR, Farnsworth GL, Bailey LL, Sauer JR (2002) Large scale wildlife monitoring studies: statistical methods for design and analysis. Environmetrics 13:1–15. https://doi.org/10.1002/env.514
Proença-Ferreira A, Ferreira C, Leitão I, Paupério J, Sabino-Marques H, Barbosa S, Lambin X, Alves PC, Beja P, Moreira F, Mira A, Pita R (2019) Drivers of survival in a small mammal of conservation concern: an assessment using extensive genetic non-invasive sampling in fragmented farmland. Biol Conserv 230:131–140. https://doi.org/10.1016/j.biocon.2018.12.021
R Core Team (2020) R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. https://www.R-project.org/
Revelle W (2015) psych: Procedures for personality and psychological research (Version 2.2.9). Evanston, IL: Northwestern University. http://CRAN.R-project.org/package=psych. Accessed 10 Oct 2022
Reynolds JH, Thompson WL, Russell B (2011) Planning for success: identifying effective and efficient survey designs for monitoring. Biol Conserv 144:1278–1284. https://doi.org/10.1016/j.biocon.2010.12.002
Rhodes JR, Jonzén N (2011) Monitoring temporal trends in spatially structured populations: How should sampling effort be allocated between space and time? Ecography 34:1040–1048. https://doi.org/10.1111/j.1600-0587.2011.06370.x
Ringvall A, Stahl G, Lamas T (1998) Guided transect sampling - a new design combining prior information and field surveying. Integrated Tools for Natural Resources Inventories in the 21st Century, Idaho, USA.
Royle JA, Young KV (2008) A hierarchical model for spatial capture-recapture data. Ecology 89:2281–2289. https://doi.org/10.1890/07-0601.1
Sabino-Marques H, Ferreira CM, Paupério P, Costa P, Barbosa S, Encarnação C, Alpizar-Jara R, Alves PC, Searle JB, Mira A, Beja P, Pita R (2018) Combining genetic non-invasive sampling with spatially explicit capture-recapture models for density estimation of a patchily distributed small mammal. Eur J Wildl Res 64:44. https://doi.org/10.1007/s10344-018-1206-x
San Miguel A (1992) Inventario de la población espanñola de Topillo de Cabrera (Microtus cabrerae Thomas, 1906). Universidad Politécnica de Madrid, Madrid
Santos SM, Simões MP, Mathias ML, Mira A (2006) Vegetation analysis in colonies of an endangered rodent, the Cabrera vole (Microtus cabrerae), in southern Portugal. Ecol Res 21:197–207. https://doi.org/10.1007/s11284-005-0104-3
Ståhl G, Ringvall A, Lamas T (2000) Guided transect sampling for assessing sparse populations. For Sci 46(1):108–115. https://doi.org/10.1093/forestscience/46.1.108
Stanley TR, Royle JA (2005) Estimating site occupancy and abundance using indirect detection indices. J Wildl Manage 69:874–883. https://doi.org/10.2193/0022-541X(2005)069[0874:ESOAAU]2.0.CO;2
Steenweg R, Hebblewhite M, Whittington J, Lukacs P, McKelvey K (2018) Sampling scales define occupancy and underlying occupancy-abundance relationships in animals. Ecology 99:172–183. https://doi.org/10.1002/ecy.2054
St-Laurent M-H, Ferron J (2008) Testing the reliability of pellet counts as an estimator of small rodent relative abundance in mature boreal forest. J Negat Results 5(1):14–22
Thompson WL (ed) (2004) Sampling rare or elusive species: concepts, designs, and techniques for estimating population parameters. Island Press, Washington, D.C
Valerio F, Ferreira E, Godinho S, Pita R, Mira A, Fernandes N, Santos SM (2020) Predicting microhabitat suitability for an endangered small mammal using Sentinel-2 data. Remote Sens 12:562. https://doi.org/10.3390/rs12030562
Valière N (2002) GIMLET: A computer program for analysing genetic individual identification data. Mol Ecol Notes 2:377–379. https://doi.org/10.1046/j.1471-8286.2002.00228.x-i2
Ventura J, López-Fuster MJ, Cabrera-Mllet M (1998) The Cabrera vole, Microtus cabrerae, in Spain: a biological and a morphometric approach. Neth J Zool 48:83–100. https://doi.org/10.1163/156854298X00237
Waits JL, Leberg PL (2000) Biases associated with population estimation using molecular tagging. Anim Conserv 3:191–199. https://doi.org/10.1111/j.1469-1795.2000.tb00103.x
Ward RJ, Griffiths RA, Wilkinson JW, Cornish N (2017) Optimising monitoring efforts for secretive snakes: a comparison of occupancy and N-mixture models for assessment of population status. Sci Rep 7(1):18074. https://doi.org/10.1038/s41598-017-18343-5
Watkins AF, McWhirter JL, King CM (2010) Variable detectability in long-term population surveys of small mammals. Eur J Wildl Res 56:261–274. https://doi.org/10.1007/s10344-009-0308-x
Wemmer C, Kunz TH, Lundie-Jenkins G, McShea WJ (1996) Mammalian sign. In: Wilson DE, Cole FR, Nichols JD, Rudran R, Foster MS (eds) Measuring and monitoring biological diversity: standard methods for mammals. Smithsonian Institution Press, Washington, D.C., USA, pp 157–176
Wilson GJ, Delahay RJ (2001) A review of methods to estimate the abundance of terrestrial carnivores using field signs and observation. Wildl Res 28:151–216. https://doi.org/10.1071/WR00033
Witmer G (2005) Wildlife population monitoring: some practical considerations. Wildl Res 32:259–263. https://doi.org/10.1071/WR04003
Woodroffe GL, Lawton JH, Davidson WL (1990) Patterns in the production of latrines by water voles (Arvicola terrestris) and their use as indices of abundance in population surveys. J Zool 220:439–445. https://doi.org/10.1111/j.1469-7998.1990.tb04317.x
Yoccoz NG, Nichols JD, Boulinier T (2001) Monitoring of biological diversity in space and time. Trends Ecol Evol 16:446–453. https://doi.org/10.1016/S0169-5347(01)02205-4
Acknowledgements
We thank all landowners who gave us permission to access their farmlands, and an anonymous reviewer for his/her careful reading of our manuscript and insightful comments and suggestions to improve it.
Funding
Open access funding provided by FCT|FCCN (b-on). This study was funded by Fundo Europeu de Desenvolvimento Regional (FEDER) through the Programa Operacional Factores de Competitividade (COMPETE) and national funds through the Portuguese Foundation for Science and Technology (FCT) within the scope of the projects ‘MateFrag’ (PTDC/BIA-BIC/6582/2014) and ‘Agrivole’ (PTDC/BIA-ECO/31728/2017). DP was supported by the FCT grant SFRH/BD/133375/2017. TM was supported by the FCT grant SFRH/BD/145156/2019. PB was supported by EDP Biodiversity Chair. JP was supported by the European Union’s Horizon 2020 research and innovation programme under project EnvMetaGen (grant agreement no 668981). RP was supported by FCT, through a research contract under the Portuguese Decree-Law nr 57/2016.
Author information
Authors and Affiliations
Contributions
Conceptualization: R. Pita, J. Paupério, X. Lambin, and P. Beja; field work: D Peralta, M. Vaz-Freire, and T. Mendes; genetic analyses: C. Ferreira and J. Paupério; statistical analyses, D. Peralta, and R. Pita; data curation: J. Paupério, and R. Pita; writing—original draft preparation: D. Peralta, and R. Pita; writing—review and editing: all authors; funding acquisition: R Pita, and J Paupério. All authors have read and agreed to the published version of the manuscript.
Corresponding author
Ethics declarations
Ethics approval
The results described in this paper were fully based on non-invasive sampling methods, which do not require approval by an Institutional Animal Care and Use Committee (IACUC) or equivalent animal ethics committee.
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Peralta, D., Vaz-Freire, T., Ferreira, C. et al. From species detection to population size indexing: the use of sign surveys for monitoring a rare and otherwise elusive small mammal. Eur J Wildl Res 69, 9 (2023). https://doi.org/10.1007/s10344-022-01634-2
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s10344-022-01634-2