Abstract
Habitat fragmentation due to anthropogenic activities is the major cause of biodiversity loss. Endemic and narrowly distributed species are the most susceptible to habitat degradation. Penstemon scariosus is one of many species whose natural habitat is vulnerable to industrialization. All varieties of P. scariosus (P. scariosus var. albifluvis, P. scariosus var. cyanomontanus, P. scariosus var. garrettii, P. scariosus var. scariosus) have small distribution ranges, but only P. scariosus var. albifluvis is being considered for listing under the Endangered Species Act. We used eight microsatellites or simple sequence repeats (SSRs) loci and two amplified fragment length polymorphism (AFLP) primer combinations to investigate the population genetic structure and diversity of P. scariosus varieties. Moreover, we compared the utility of the two marker systems in conservation genetics and estimated an appropriate sample size in population genetic studies. Genetic differentiation among populations based on Fst ranged from low to moderate (Fst = 0.056–0.157) and from moderate to high when estimated with Des (Des = 0.15–0.32). Also, AMOVA analysis shows that most of the genetic variation is within populations. Inbreeding coefficients (Fis) were high in all varieties (0.20–0.56). The Bayesian analysis, STRUCTURE, identified three clusters from SSR data and four clusters from AFLPs. Clusters were not consistent between marker systems and did not represent the current taxonomy. MEMGENE revealed that a high proportion of the genetic variation is due to geographic distance (R2 = 0.38, P = 0.001). Comparing the genetic measurements from AFLPs and SSRs, we found that AFLP results were more accurate than SSR results across sample size when populations were larger than 25 individuals. As sample size decreases, the estimates become less stable in both AFLP and SSR datasets. Finally, this study provides insight into the population genetic structure of these varieties, which could be used in conservation efforts.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Anthropogenic activities are the main cause of biodiversity loss today (Pimm and Raven 2000; Ceballos et al. 2015). In the United States, most endangered plants (81%) are threatened by habitat loss and degradation. The expansion of agricultural boundaries (excluding pasturing) is the leading cause of habitat destruction in the United States (Wilcove et al. 1998). As a consequence, 33% of endangered plants are affected by livestock grazing. Likewise, 133 species in the United States are threatened due to activities linked to mining for oil and gas (Wilcove et al. 1998). The state of Utah has the largest mining excavation in the world with the second largest copper producer in the United States (Lee 2016). Also, there are 42 Utah species listed under the Endangered Species Act (ESA), and 60% of them are plants (U.S. Fish and Wildlife 2017).
Penstemon, with ca. 275 species, is the largest genus of plants endemic to North America (Wolfe et al. 2006). Utah harbors the largest number of native and endemic species in the genus (Holmgren 1984; Nold 1999; Wolfe et al. 2006). There are three species of Penstemon listed under the ESA [P. debilis, P. haydenii, and P. penlandii (U.S. Fish and Wildlife 2017)]. In 2013, the U.S. Fish and Wildlife Services (USFWS) introduced a regulation designating critical habitat for P. grahamii and P. scariosus var. albifluvis to prevent environmental destruction in the areas where these two taxa occur (USFWS 2013). However, in 2014, the proposed rule was withdrawn and as a consequence, habitat conservation for these two taxa was returned to private organizations and landowners. Currently, the USFWS reinstated the 2013 proposed rule for both species following a court order (Rocky Mtn. Wild, et al. v. Walsh 2016). In order to develop conservation strategies for these species, an understanding of the genetic diversity and geographical distribution is needed.
Population genetic studies can provide an important perspective for conservation. According to several authors, many threatened species have low genetic diversity and high inbreeding coefficients (Saccheri et al. 1998; Spielman et al. 2004; Frankham et al. 2010; Rodríguez-Peña et al. 2014a). These consequences are more acute in small populations found in fragmented habitats. Fragmentation of populations, regardless of their sizes, will reduce gene flow (Aguilar et al. 2008), decrease genetic diversity (Lowe et al. 2005), and magnify the impact of genetic drift (Young et al. 1996). Even though population genetic studies are recognized to be relevant in addressing conservation strategies (Ouborg et al. 2006; Frankham et al. 2010), few conservation initiatives consider genetics as part of the resource management process (Mijangos et al. 2015). Additionally, budgetary constraints can limit not only the number of samples that can be included in a study, but also the feasibility of a genetic survey. Conservationists need affordable tools that allow for fast and easy implementation of population genetics in conservation strategies.
The ideal sample size for population genetic studies is controversial (discussed in Hale et al. 2012; Kalinowski 2005). Intrinsic aspects of populations, such as inbreeding and genetic diversity, can influence how many samples are required to define a representative estimator of population genetic parameters (Bashalkhanov et al. 2009). For example, when inbreeding is high and populations are small, few individuals will be sufficient to capture most alleles in the population. In contrast, when genetic diversity is large and inbreeding coefficients are low, a much larger sample size is needed. However, many authors argue that 20–30 individuals are sufficient to determine population diversity and differentiation (Hale et al. 2012; Kalinowski 2005; Sinclair and Hobbs 2009).
Genetic parameters are affected by several life history traits. In particular, breeding system is the factor that influences the most demographic estimates (Duminil et al. 2007; Hamrick and Godt 1996). In a meta-analysis of random amplified polymorphic DNA (RAPD) in plants, Nybom and Bartish (2000) found that outcrossing species have higher within-population genetic diversity, and lower between-population genetic diversity than selfing taxa.
Extrinsic factors such as the number of loci and type of marker used could also affect estimates of population genetic parameters. Cavers et al. (2005) determined the optimal sampling strategy in order to obtain reliable spatial genetic structure (SGS) estimates in tree populations. The authors reported that a much larger number of amplified fragment length polymorphism (AFLP) loci and individuals are needed to achieve the same results in comparison with microsatellites. That study, however, only included sample sizes that ranged from 50 to 200 and loci numbers ranging from 1 to 100. Cavers et al. (2005) also found that a dataset of 150 individuals and 100 loci (two primers) for AFLP and 100 individuals and 10 loci for simple sequence repeats (SSRs) are sufficient to obtain reliable estimates of SGS parameters. The authors did not evaluate the effect of more than 100 loci and less than 50 individuals. In contrast to Cavers et al. (2005), Jump and Peñuelas (2007) found that 150–200 individuals and 100–150 AFLP loci were sufficient to accurately determine SGS in the tree Fagus sylvatica, while 150–200 individuals and six SSR loci were not.
The nature of the marker (AFLP or SSR) could have an effect on population genetic estimates. Lower levels of genetic differentiation (Fst) are expected with SSRs due to the higher mutation rates of those markers (Gaudeul et al. 2004; Maguire et al. 2002) while higher degrees of genetic differentiation are expected with AFLP, because they are more likely to be linked with loci under selection (Jump and Penuelas 2007). Additionally, calculations of some estimates are also limited by the type of marker. Expected heterozygosity can only range from 0 to 0.50 when calculated with AFLP data (Gaudeul et al. 2004) because AFLP loci can be either absent or present.
The choice of genetic marker can also affect inferences of genetic diversity in some plant species. Recent studies have applied SSRs (Rodríguez-Peña et al. 2014b; Wolfe et al. 2016), ALFPs (Waycott and Barnes 2001; El-Esawi et al. 2016), SNPs (Cahill and Levinton 2016; Kaiser et al. 2017), and other genetic markers. Variability in the marker loci is important, but the cost of conducting a genetic survey is often a limiting factor.
Microsatellites, or simple sequence repeats (SSRs), are thought to be neutral and highly variable molecular markers (Ellegren 2004) widely used to assess population genetic structure and diversity (Sunnucks 2000; Jarne and Lagoda 1996; Allendorf et al. 2010; Rodríguez-Peña et al. 2014a). They are also widely used to investigate demographic history (Pritchard et al. 1999; Castellanos-Morales et al. 2016) and delimit species boundaries (Caddah et al. 2013; Rodríguez-Peña et al. 2014b). Microsatellites allow for the calculation of expected and observed heterozygosity, and the inbreeding coefficient (Fis) (Morgante and Olivieri 1993; Putman and Carbone 2014). The microsatellite technique has a disadvantage in that prior information of DNA sequence flanking repeat motifs is required (Sønstebø et al. 2007). Also, SSRs can be relatively expensive to design, may have low transferability across species (Garcia et al. 2004), and often involve multiple steps to identify reliable markers (Kalia et al. 2011; Kaiser et al. 2017). However, microsatellite primers designed for Penstemon have high transferability across species (Dockter et al. 2013; Wolfe et al. 2016). Additionally, a new high-throughput sequencing genotype pipeline has been developed to reduce labor and cost of SSRs analysis in comparison with capillary electrophoresis (De Barba et al. 2017). SSRs are highly polymorphic (Powell et al. 1996), are fairly abundant in the genome (Agarwal et al. 2008), and allow the differentiation between heterozygous and homozygous individuals (Garcia et al. 2004).
AFLP markers are often used in plant genetic studies to investigate population structure and diversity (Waycott and Barnes 2001; Campbell et al. 2003), to elucidate complex taxonomic relationships (Riek et al. 2013), and in DNA fingerprinting studies (Vos et al. 1995; Miggiano et al. 2005; Fajardo et al. 2013). The biggest benefit of using AFLPs is that no previous knowledge of DNA sequences is required (Sønstebø et al. 2007). Additionally, AFLPs are highly polymorphic and replicable, and they yield many loci with few primer combinations. The AFLP technique is fast, robust, and reliable (Meudt and Clarke 2007; Costa et al. 2016). The biggest disadvantage of AFLPs is that they are dominant markers, which does not allow the exact calculation of observed heterozygosity and the inbreeding coefficient (Garcia et al. 2004). However, calculation of expected heterozygosity (Peakall and Smouse 2012) and estimates of the inbreeding coefficient (Chybicki et al. 2011) are available.
In this study, we investigate the genetic diversity and differentiation of the P. scariosus species complex. The overarching goal of this research is to provide conservation agencies with tools to create comprehensive conservation strategies for P. scariosus varieties. We first aim to evaluate the minimum number of individuals required to accurately calculate basic genetic statistics. Second, we compare the fidelity of each marker system (AFLP and microsatellite) across sampling subsets.
Methods
Study species and sample collection
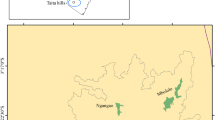
Penstemon scariosus (Plantaginaceae) is a complex of taxa comprised of four varieties, P. sc. var. albifluvis (England) N. Holmgren, P. sc. var. cyanomontanus Neese, P. sc. var. garrettii (Pennell) N. Holmgren, and P. sc. var. scariosus Pennell. All varieties are perennial herbs/subshrubs with purple-lavender flowers, mostly pollinated by insects (Online Resource 1, Figure S1). They are distributed in the northeastern region of Utah, the northwest border area of Colorado, and the southwest border area of Wyoming (Fig. 1). Morphologically, these four varieties are very similar, and are mostly differentiated by their geographic and edaphic distribution. Penstemon scariosus var. albifluvis is endemic to oil shale (England 1982) found in Uintah Co., Utah and adjacent Rio Blanco Co., Colorado (Holmgren 1984). Penstemon scariosus var. cyanomontanus occurs in sandstone (Neese 1986) and quartzite substrate on the Diamond Mt., Mosby Mt., summit of Blue Mt., and adjacent Colorado (Goodrich and Neese 1986). Penstemon scariosus var. garrettii grows in travertine (Pennell 1920) and calcareous rocks (Goodrich and Neese 1986) from the eastern side of the Wasatch Range to Uinta Mts., Blue Mts., and Tavaputs plateaus. Finally, P. scariosus var. scariosus inhabits the Wasatch, Fish Lake, and the northern Aquarius plateaus (Holmgren 1984).
Distribution map of P. scariosus varieties. Samples used in this study represent the complete distribution of the varieties. The color scheme indicates the four varieties as follows: black P. sc. var. albifluvis, blue P. sc. var. cyanomontanus, pink P. sc. var. garrettii, and green P. sc. var. scariosus. (Color figure online)
We sampled from 59 sites, 1–23 individuals per site, across the entire known range of the P. scariosus complex, and 561 individuals across varieties (Table 1; Fig. 1). All individuals were included in the AFLP survey. However, only 421 individuals were sampled for SSRs due to budgetary constraints. Our initial sampling included sites geographically close to each other. We trimmed the original dataset from 561 to 421 individuals by excluding geographically close samples. Two to three young healthy leaves were collected from independent samples at each collection site (Online Resource 1, Table S1) and placed in individually labeled “extra slim” tea filter bags (Finum.com, Riensch & Held GmbH & Co. KG, Hamburg, Germany). We immediately placed eight plant samples in an airtight plastic bag, with no less than 100 g of dry silica gel. Once returned to the laboratory, the samples were immediately stored at − 80 °C until the samples could be completely lyophilized. After lyophilized, all samples were stored in sealed plastic bags with silica gel at − 20 °C until DNA extraction. DNA was extracted from dried leaf tissue using the method detailed by Todd and Vodkin (1996).
SSR and AFLP genotyping
Eight previously designed microsatellite nuclear loci (Kramer and Fant 2007; Dockter et al. 2013; Anderson et al. 2016) were used to assess population genetic estimates for the P. scariosus species complex (Online Resource 1, Table S3). Microsatellites were amplified following the specification in Anderson et al. (2016). Samples were run on an ABI 3730xl Genetic Analyzer (Applied Biosystems, Foster City, CA, USA) with Gene Scan 500 ROX Size Standard (Applied Biosystems) at the Brigham Young University DNA Sequencing Center (Provo, Utah).
AFLP markers were designed using the AFLP Plant Mapping Kit from Applied Biosystems (Online Resource 1, Table S2). Out of the 66 resulting primer combinations, we chose the two most diverse. The AFLP protocol followed Vos et al. (1995), with modifications. In particular, the first step included restriction with MseI and EcoRI and subsequent ligation with T4 DNA ligase (New England BioLabs, Ipswich, MA, USA), made in the lab Eco-Adaptor, Mse-Adaptor (Online Resource 1, Table S2), and Purified BSA from BioLabs (2 h at 37 °C in a thermocycler and then overnight at room temperature). In the second step, pre-selective amplification, we used the digested and ligated product from the previous step and added DreamTaq Green polymerase (Thermo Fisher Scientific, Waltham, MA, USA), dNTPs (Applied Biosystems), Eco-primer, and Mse-primer (Online Resource 1, Table S2). The final step, selective amplification, was carried out with DreamTaq Green polymerase, dNTPs, and selected primers (each primer pair had one fluorescent-labeled primer, Online Resource 1, Table S2). AFLP samples were run on an ABI Genetic Analyzer 3100 after denaturation with HiDi formamide (Applied Biosystems).
Data analyses
Peak Scanner v.1.0 (Applied Biosystems) was used to visualize and score the SSR fragments, while GeneMapper 3.1 (Life Technologies) was used to score AFLP alleles. MICRO-CHECKER 2.2.3 (Van Oosterhout et al. 2004) was used to test the presence of null alleles, large allele dropout, genotyping errors, and stuttering for SSRs. We used GenAlEx v. 6.501 (Peakall and Smouse 2012) to record genotypes for both SSRs and AFLPs, and calculated basic genetic statistics, including number of private alleles (np), average number of alleles per locus (A), percentage of polymorphic loci (P), number of identical genotype pairs (nig), unbiased expected heterozygosity (uHe), expected heterozygosity (He), and observed heterozygosity (Ho), for SSRs only.
For microsatellite data, we used Arlequin v. 3.5 (Excoffier and Lischer 2010) to produce basic genetic statistics, such as the inbreeding coefficient (Fis), genetic differentiation among pairs of populations [Fst as formulated by Wright (1951), and modified by Weir and Cockerham (1984)], percentage of paired loci showing significant linkage disequilibrium (LDL), number of loci that deviate significantly from Hardy–Weinberg equilibrium (nds), the number of migrants per generation (Nm), and an AMOVA analysis. We used the program SMOGD (Crawford et al. 2010) to obtain the differentiation estimate (Dest) for microsatellite data as formulated by Jost (2008). This estimator is thought to yield a more reliable measure of genetic differentiation when the populations are small. The program GenePop v. 007 (Rousset 2008) was used to perform the U-test for heterozygosity deficiency for microsatellite data using 10,000 Markov Chain Monte Carlo iterations for each population (Guo and Thompson 1992). The Nei’s genetic distances and genetic differentiation (PhiPT) were calculated in GenAlEx v. 6.501 (Peakall and Smouse 2012) for both AFLP and SSR datasets. The program I4A, inbreeding for AFLPs, (Chybicki et al. 2011) was used to estimate the inbreeding coefficient (Fis) for each variety. Estimates were calculated at three different values (0.1, 1, and 5) for each parameter, alpha and beta (Oleksa et al. 2013), using 10,000 rejected steps and 50,000 sampling steps. By computing different values of alpha and beta, we assess how much the inbreeding coefficients change when the shape parameters change. The accuracy of this estimate is determined by comparing the similarities between the log-likelihood values among the different B-distribution parameters. High similarity implies high accuracy.
STRUCTURE V2.3.4 (Pritchard et al. 2000) was used to infer the number of clusters. This method assigns individuals to groups based on the allele frequency, using the Monte Carlo Markov Chain (MCMC) approach. The admixture model and correlated allele frequencies were used, as recommended by Falush et al. (2003), with 10,000 burn-in periods and 50,000 MCMC repetitions after burn-in (Evanno et al. 2005). Values of K ranged from 1 to 10, with 20 independent replications. To obtain the “true” value of K, the delta method of Evanno et al. (2005), as implemented in STRUCTURE HARVESTER (Earl and vonHoldt 2012), was used. The replicates from the best K-value were aligned with the software CLUMPP (Jakobsson and Rosenberg 2007), and a Q-matrix was created. The results from CLUMPP 1.1.2 were visualized with the program DISTRUCT 1.1 (Rosenberg 2004). The R package MEMGENE (Galpern et al. 2014) was used to detect spatial patterns of genetic structure among all individuals. MEMGENE is used to detect cryptic spatial genetic patterns that other approaches, such as the Mantel test, are unable to detect. A genetic distance matrix based on Cavalli-Sforza and Edwards (1967) was constructed in the program POPULATIONS (Langella 2007) for SSRs, and a matrix based on Nei’s genetic distance was constructed in AFLP-SURV 1.0 (Vekemans et al. 2002) for AFLP. From these matrices, we inferred similarities across varieties using the program NEIGHBOR from PHYLIP 3.6 (Felsenstein 2005). The resulting tree was visualized in TREEVIEW 1.6.6 (Page 1996).
Comparison
For the comparison within AFLP, we created a master dataset that had the same individuals as in the SSR dataset (421). We were interested in studying the relative performance of different sample sizes in comparison to the full dataset within the same marker system (AFLP and SSR). Subsets of the full dataset were constructed as follows: we randomly resampled from the full AFLP dataset multiple times, using both a fixed proportion and a fixed number of individuals. Specifically, we resampled using nine different fixed proportions of individuals (10%, 20%, 30%, 40%, 50%, 60%, 70%, 80%, 90%) and six different fixed numbers of individuals (5, 10, 15, 20, 25, 30), creating 15 subsets. We repeated this process for the SSR dataset. For each subset, we replicated the resampling five times. Every subset contained a proportional representation of individuals from the four varieties (Table 1). For example, the subset “10%” had 23, 6, 19, and 9 individuals from varieties P. sc. var. albifluvis, P. sc. var. cyanomontanus, P. sc. var. garrettii, and P. sc. var. scariosus, respectively, for a total of 57 individuals. For every subset, we obtained basic genetic statistics (He, uHe, A, P, np, nig), AMOVA, Fst, adjusted R-squared (from MEMGENE results), and paired population-wise genetic distance and differentiation. We compared the values obtained from each subset to the full dataset values to determine significant differences at P < 0.05 with individual paired t-tests. The relative accuracy of each subset depended on the differences between the values obtained from the subset and those from the full dataset. Therefore, a smaller difference between the subset and the full dataset implies high accuracy of the value estimated from the subset. For example, the mean R-squared value obtained from the iterations of the 10% and 90% subsets was 0.27 and 0.36, respectively, while the value obtained from the full dataset was 0.38. After performing a t-test, we determined that the R-squared values from the 10% subset were significantly different from those obtained when using the full dataset. However, the 90% subset was not statistically different from the full dataset. This comparison also allowed us to determine the consistency of each marker. The marker is consistent when the sample size and the accuracy of the estimates are positively correlated.
To make results comparable between markers, we divided the standard deviation by the mean value to adjust the estimates. The standard deviation and the mean of every genetic result (AMOVA, genetic distance, etc.) were calculated from each subset. The standard deviation measures the spread of the genetic results in every subset individually. A larger standard deviation implies a larger spread of the results in that particular subset. A paired t-test was performed on the adjusted standard deviation of every subset between AFLPs and microsatellites. The objective of this comparison was to determine whether the results from each marker type were significantly different at a p-value less than 0.05.
Results
Genetic diversity and differentiation: microsatellite
The microsatellite dataset, obtained from eight SSR primers (Online Resource 1, Table S3), included 421 individuals from the four varieties of P. scariosus (Table 1) and had 0.4% missing data. All missing data were from two of the loci, PEN04 and PEN23. The complete SSRs dataset included 142 alleles from eight loci (PS084 (13), PS082 (15), Pen04 (18), PS080 (23), Pen23 (25), PS014 (15), PS016 (20), PS048 (13)). Genetic statistics for all SSR markers are summarized in Table 2. Analyses with MICRO-CHECKER indicated homozygote excess in all loci, but no evidence for large allele dropout. There was an average of 11 alleles per locus, where P. sc. var. garrettii had the largest number of alleles per locus (15). Observed heterozygosity (Table 2), on average, was moderate (0.560). In this respect, P. sc. var. scariosus had the least genetic diversity (0.460), while P. sc. var. cyanomontanus had the largest (0.633). The inbreeding coefficient (Fis) ranged from 0.200 to 0.370. Contrary to the expectation that smaller populations have less genetic diversity, P. sc. var. scariosus had the largest inbreeding coefficient and the least genetic diversity, while P. sc. var. cyanomontanus, with the smallest number of individuals sampled, had the largest amount of genetic diversity and the smallest inbreeding coefficient. There were no identical genotype pairs in any of the varieties. The mean number of migrants per generation was high (Table 3). In particular, the highest number of migrants was between P. sc. var. garrettii and P. sc. var. scariosus, and the smallest was between P. sc. var. albifluvis and P. sc. var. scariosus (8.916 and 3.179, respectively). Genetic differentiation between pairs of varieties was low to moderate (Table 3). The largest genetic differentiation (Fst) was between P. sc. var. scariosus and P. sc. var. albifluvis (0.157), and the lowest genetic differentiation was between P. sc. var. garrettii and var. scariosus (0.056). As expected, Des genetic differentiation was larger than Fst (Table 3, Online Resource 1, Table S4), but the Des values were proportional to Fst. Penstemon scariosus var. scariosus P. sc. var. albifluvis had the largest genetic differentiation (0.320), and P. sc. var. garrettii and P. sc. var. scariosus had the lowest (0.150). The Nei’s genetic distance (Online Resource 1, Table S5) was consistent with the number of migrants per generation, and varieties with the largest genetic distances had the smallest number of migrants per generation, and vice versa. The AMOVA analysis indicated that 67% of the genetic variation is within individuals, whereas only 9% is among varieties and 24% among individuals.
Genetic diversity and differentiation: AFLP
The two primers selected yielded 490 loci across 561 individuals and 0% missing data. AFLP polymorphism was different between the varieties and ranged from 39% (P. sc. var. cyanomontanus) to 74% (P. sc. var. garrettii). Genetic statistics are summarized in Table 4. The mean number of alleles per locus was 0.510. The lowest number of alleles per locus was in P. sc. var. scariosus (0.044), while the largest was in P. sc. var. albifluvis (1.155). The expected heterozygosity varied from 0.080 to 0.650. The inbreeding coefficient was high in all varieties (Table 4). The largest inbreeding coefficient was in P. sc. var. scariosus (0.560), while the lowest was in P. sc. var. garrettii (0.300). The log-likelihood values were highly similar within varieties, which means that the estimates of the inbreeding coefficient are stable regardless of the values used for the shape parameters (Table 5). The variety P. sc. var. garrettii had the highest number of private alleles (89), and P. sc. var. cyanomontanus had the lowest (14), which is not proportional to the average number of alleles in each variety (Table 4). The Nei’s genetic distance (Online Resource 1, Table S6) was in accordance with the genetic differentiation. The smallest genetic distance and genetic differentiation were between P. sc. var. albifluvis and P. sc. var. cyanomontanus (0.005 and 0.100, respectively). The largest genetic distance (0.015) and genetic differentiation (0.230) were between P. sc. var. cyanomontanus and P. sc. var. garrettii. The AMOVA analysis showed that 81% of the variation is within varieties and 19% among varieties.
Genetic structure: microsatellite
MEMGENE results (Fig. 2) showed that a moderate proportion of the genetic variation is explained by the spatial pattern of distribution (R2 = 0.28, P = 0.001). The spatial pattern detected in genetic distance data indicated that P. sc. var. cyanomontanus, P. sc. var. garrettii, and P. sc. var. scariosus are the most genetically similar. Consequently, P. sc. var. albifluvis formed an isolated group. Graphical representations of these results are shown in Fig. 2, where the size and the color of the circles represent genetic similarity, and the x-axis and y-axis represent longitude and latitude, respectively. Using the Delta-K method, as implemented in STRUCTURE HARVESTER, we found that K = 2 (Online Resource 1, Figure S2) was the most likely value followed by K = 3 (Online Resource 1, Figure S3). When K = 2, P. sc. var. albifluvis formed an isolated group with almost no admixture, leaving P. sc. var. cyanomontanus, P. sc. var. garrettii, and P. sc. var. scariosus to form the second cluster. When K = 3, P. sc. var. albifluvis still forms a distinct group, but in this case, P. sc. var. cyanomontanus and P. sc. var. scariosus also form distinct groups. Finally, P. sc. var. garrettii is highly admixed with the other three varieties. The neighbor-joining tree (Fig. 3) shows two separate clusters. The first cluster includes varieties P. sc. var. albifluvis and P. sc. var. cyanomontanus, while the second is formed by varieties P. sc. var. garrettii and P. sc. var. scariosus. Penstemon scariosus var. scariosus was the most genetically distant, as indicated by the long branch length.
MEMGENE plots for the first axis based on pairwise genetic and geographic distance. The size and the color of the circle represent genetic similarity between individuals, and the x-axis and y-axis represent longitude and latitude, respectively. The plot on the left is based on eight microsatellite loci, and the plot on the right is based on 490 AFLP loci
Neighbor-Joining tree based on Nei’s genetic distances between populations. The tree on the left is based on eight microsatellite loci, and the tree on the right is based on 490 AFLP loci. Both trees depict the same relationship between the varieties. Penstemon scariosus var. albifluvis and P. sc. var. cyanomontanus form one cluster, and P. sc. var. garrettii and P. sc. var. scariosus form another
Genetic structure: AFLP
The neighbor-joining tree (Fig. 3) yielded two clusters: one with P. sc. var. albifluvis and P. sc. var. cyanomontanus, and the other with P. sc. var. scariosus and P. sc. var. garrettii. Penstemon scariosus var. cyanomontanus had the longest branch length, which means this variety is genetically more distant from the other varieties. We visualized the result of STRUCTURE after running STRUCTURE HARVESTER, CLUMPP, and DISTRUCT. Even though the “best” value of K was 4 (Online Resource 1, Figure S4), there is not a clear separation of the varieties; hence, all varieties are highly admixed. The results from MEMGENE (Fig. 2), which estimates what proportion of genetic variation is due to geographic distance, were consistent with our structure analyses, where P. sc. var. albifluvis and P. sc. var. cyanomontanus form a group. Specifically, MEMGENE results indicated that there is a strong spatial correlation between individuals (R2 = 0.38, P = 0.001). Penstemon scariosus var. garrettii and P. sc. var. scariosus were clustered together, while P. sc. var. albifluvis and P. sc. var. cyanomontanus formed another group.
Comparisons between different sample sizes and between AFLPs and SSRs
The first level of comparison was among subsets of marker types, to assess how much the sample size can decrease before the population genetic estimators are significantly different from those of the largest sample size. Since it is not realistic to compare subsets that exceed the budget of most projects, we focused the comparisons of the subsets of 5–30 individuals, and 10–30% of the total sample size (see details in “Methods”).
Thirty ANOVA tests (1050 pairwise comparisons) were performed independently to compare subsets from each AFLPs and SSRs, individually. For the AFLP data subsets, we found that the larger the sample size, the closest the estimates were to those calculated from the full dataset (Online Resource 1, Figures S5 and S6). As a general trend, larger subsets yielded more accurate estimates. The paired t-tests revealed that the subset of 30 individuals was more accurate than that of 25 individuals twice, and the subset of 25 individuals was more precise than the subset of 20 individuals six times. The comparison made within the SSRs subsets yielded similar results. Twice was the subset of 30 individuals more accurate than that of 25 individuals. Also, the subset of 25 individuals was more accurate than the subset of 20 individuals three times. The subsets 10% and 15, 10, and 5 individuals were less accurate compared to larger ones in both datasets (AFLP and SSR).
The second level of comparison was between markers (AFLP and SSR). We compared 30 measurements, including paired population genetic distance using a paired t-test each time. Less than half (13) of the t-tests were statistically significant (P < 0.05) when all 15 subsets were in the analysis. It is important to highlight that there were no significant differences between AMOVA statistics (Online Resource 1, Figures S7 and S8) and adjusted R-squared (MEMGENE analysis, Online Resource 1, Figures S9 and S10) calculated from different markers. In contrast, when the subsets 5–25 individuals were excluded from the analysis, in only two comparisons was the difference between AFLPs and SSRs significant. In these two cases, each marker type performed the best once.
Discussion
Genetic diversity and differentiation
Genetic diversity, as measured by observed heterozygosity and the average number of alleles per locus, was higher than expected despite the varieties having a small number of individuals throughout their distribution range. Currently, the only variety with an estimate of population size is P. sc. var. albifluvis. In its most recent survey, the U.S. Dept. of Interior - Fish and Wildlife Service (USFWS 2014) estimated the total population size of this variety to be 12,215 individuals. Genetic diversity within the P. scariosus species complex (Table 2) is similar to that in other species of Penstemon with conservation concerns (Wolfe et al. 2014, 2016). For example, the estimated number of individuals of P. albomarginatus ranges from 100,000 to 200,000, and this species has fewer alleles per locus, slightly higher observed heterozygosity, and lower inbreeding (A = 7.89, Ho = 0.62, Fis = 0.16; Wolfe et al. 2016). Penstemon debilis, on the other hand, has a smaller estimated number of individuals (4100) than the P. scariosus species complex, with fewer alleles per locus, lower observed heterozygosity, and a very similar inbreeding coefficient [A = 4.16, Ho = 0.43, Fis = 0.23; (Wolfe et al. 2014)]. Despite this apparent high level of genetic diversity, the inbreeding coefficients we report are high, even in P. sc. var. garrettii, which has the lowest Fis (Table 4) and the second largest number of individuals (Table 1). Penstemon scariosus var. scariosus, with the second smallest number of individuals, has the highest inbreeding coefficient, lower than average alleles per locus, and the lowest observed heterozygosity. Additionally, it is located at the southernmost point of the P. scariosus species complex distribution range (Fig. 2) and is isolated from the rest of the varieties. In contrast, P. sc. var. cyanomontanus, has one of the lowest inbreeding values, the lowest average alleles per locus, but the highest observed heterozygosity, and the smallest number of individuals. Furthermore, P. sc. var. cyanomontanus occurs in close proximity to P. sc. var. albifluvis and P. sc. var. garrettii. Thus, isolated groups of individuals are more inbred than small, marginally sympatric ones.
Patterns of genetic differentiation (Table 3, Online Resource 1, Tables S4 and S6) are consistent with results from the program STRUCTURE. However, the STRUCTURE results from SSRs (Online Resource 1, Figures S2 and S3) indicated that P. sc. var. albifluvis formed an isolated group, while STRUCTURE results from ALFPs (Online Resource 1, Figure S4) showed that all varieties have some level of admixture. Recently, Puechmaille (2016) demonstrated that, when sample sizes are unequal, the program STRUCTURE tends to group populations/sites that have a small number of individuals into larger groups (Puechmaille 2016). This bias affects both AFLP and SSR datasets. Moreover, STRUCTURE is sensitive to high levels of linkage disequilibrium among pairs of loci (Falush et al. 2003). Arlequin results from SSRs (Table 2) show that an average of 56% of paired loci are in linkage disequilibrium, with the highest values for P. sc. var. garrettii (84%), and the lowest for P. sc. var. cyanomontanus (21%). STRUCTURE results from SSRs could be less reliable than those from AFLPs in this study. The mismatch between results from different markers could also indicate taxonomic discrepancy. We used the traditional taxonomical delimitation of the varieties, which is mostly based on geographic distribution. The results from this paper suggest that there might be some taxonomic incongruence in the current delimitation of the varieties. Future work will focus on the phylogenetic reconstruction of P. scariosus species complex and closely related species.
We can infer from our results that geographic distance plays an important role in limiting gene flow in the P. scariosus species complex. This has enormous implications for conservation. For example, in the only breeding system study for P. scariosus, Lewinsohn and Tepedino (2007) found that P. sc. var. albifluvis has a mixed mating system, where fruits are produced by outcrossing and by selfing when pollinators are absent. The same study showed that currently, pollinator frequency maximizes seed production. This means that in the absence of pollen from other relatively close individuals, plants will self-pollinate and, as a consequence, genetic variation could decrease, genetic differentiation is likely to increase, and populations will become more sensitive to drift-based threats (Ellstrand 1992). Experimental evidence showed that loss of genetic variation is associated with inbreeding depression and an increased risk of extinction in Clarkia pulchella (Newman and Pilson 1997). More generally, Brook et al. (2002) compared the median time to extinction of 20 threatened species using computer simulation, and found that inbreeding depression reduces the time to extinction by 25–30%.
According to our survey, Penstemon scariosus varieties have small areas of distribution (Fig. 1). Our results show that most of the varieties have high inbreeding coefficients and low genetic diversity. These estimates are consistent with those in similar studies of Penstemon species with small population sizes, and are proportional to those of species with larger populations (Wolfe et al. 2014). Population size is one of the most important factors influencing extinction risk. Small populations are more susceptible to inbreeding depression, genetic drift, and loss of genetic diversity, relative to large ones (Hilfiker et al. 2004; Frankham et al. 2010). Genetic diversity is reduced in all populations of P. scariosus in comparison to other species of Penstemon with populations of similar sizes and natural history (Wolfe et al. 2014, 2016). The loss of genetic diversity can potentially reduce a species’ ability to adapt to new environmental conditions, and, as a consequence, it could increase the risk of extinction (Helm et al. 2009). According to Lynch et al. (1995), who developed theoretical models to assess extinction risk, populations progressively accumulate deleterious mutations as the number of individuals is reduced. They emphasize that just 12 generations are required to drive a population of fewer than 100 individuals to extinction.
In other plant species, similar patterns of genetic differentiation have been observed. For example, in an AFLP-based study of Geranium pratenses, which is a perennial outcrossing species with an estimated number of individuals per population ranging from eight to 42,000, Michalski and Durka (2012) found lower genetic diversity (He = 0.15) but a lower inbreeding coefficient (Fis = 0.21) than in P. scariosus on average. The authors also indicated that isolation by distance due to gene flow is responsible for the genetic differentiation observed between populations within 10 km, but at larger distances, genetic drift influences the observed pattern the most. Moderate to high levels of genetic differentiation (0.059–0.572) was found in Magnolia ashei, a species with a mixed mating system. The authors attribute the high levels of genetic differentiation to low gene flow induced by anthropization (Von Kohn et al. 2018).
AFLPs versus SSRs
It is difficult to determine the best sample size for all population genetic studies. Some authors have argued that 25–30 individuals per population are sufficient to accurately determine allele frequencies (Hale et al. 2012), while others argue that 20–100 individuals are required to assess genetic structure depending on Fst values (Kalinowski 2005; Sinclair and Hobbs 2009). However, assessing 30 individuals per population is challenging when many populations must be included in the analysis and the budget is limited. Our results suggest that 25 individuals per population is an adequate sample size if using AFLP loci (Online Resource 1, Figures S5, S7, and S9). When SSRs are implemented, 20 individuals is a more appropriate sample size (Online Resource 1, Figures S6, S8, and S10). Other authors have indicated that 20–30 is an adequate sample size in a population genetic study (Hale et al. 2012; Kalinowski 2005; Sinclair and Hobbs 2009), which is similar to what we found. Additionally, certain measurements are more sensitive to sample size variation than others. The average number of alleles per locus (Online Resource 1, Figures S5 and S6), and the number of private alleles (data not shown) changed dramatically from one sample size to another. However, AMOVA results were consistent across sample sizes (Online Resource 1, Figures S7 and S8).
For the second level of comparison between AFLPs and microsatellites, the adjusted R-squared values (from MEMGENE) for microsatellites were highly variable (Online Resource 1, Figure S10). In particular, results were inconsistent across the subsets. In most cases, when the sample size increases, the accuracy of the estimates also increases. However, in the case of adjusted R-squared analysis from the microsatellite dataset, values fluctuated inconsistently, as the number of individuals in the subsets increases. On the other hand, adjusted R-squared values exhibit the opposite pattern when calculated from AFLP data (Online Resource 1, Figure S9). In most cases, as the number of individuals in the subset dataset increases, the accuracy of the measure also increases, reaching stability at 20%. This was a major difference between AFLPs and microsatellites. However, if more SSR markers had been utilized, these differences may have been reduced. Some authors suggest that 5–10 SSR loci and 150–100 individuals, respectively, are sufficient to estimate spatial genetic structure (Cavers et al. 2005). In two cases, our SSRs dataset did not satisfy the minimum, as P. sc. var. cyanomontanus has 32 individuals and P. sc. var. scariosus has 65.
Increasing the sample size in a study of endangered species could be difficult, and sometimes impossible. Collecting 20–30 individuals of a rare species can be challenging, especially when populations are small. To increase sample size and maximize sampling efforts, some researchers use landscape similarities analysis and environmental niche modeling (de Siqueira et al. 2009; Guisan et al. 2006; Lomba et al. 2010). In addition to augmenting the number of samples, scientists can also increase the number of loci (De Barba et al. 2017; Vos et al. 1995).
Based on the life history traits of P. scariosus and its current estimated population size (at least 12,215 individuals), we believe our results are transferable to other species with conservation concerns. Outcrossing species, like P. scariosus, have more genetic diversity and less genetic differentiation than selfing species; therefore, 20–25 individuals should be enough to compute estimates of population genetics for species that naturally harbor less genetic diversity such as species with mixed mating and selfing system.
In summary, AFLP and SSR provide accurate estimates of population genetic parameters. Both have intrinsic advantages and disadvantages. However, when there is no previous knowledge of the target species genome and the goal is to inform conservation decisions, AFLP markers would be an excellent economical choice.
References
Agarwal M, Shrivastava N, Padh H (2008) Advances in molecular marker techniques and their applications in plant sciences. Plant Cell Rep 27:617–631
Aguilar R, Quesada M, Ashworth L et al (2008) Genetic consequences of habitat fragmentation in plant populations: susceptible signals in plant traits and methodological approaches. Mol Ecol 17:5177–5188
Allendorf FW, Hohenlohe PA, Luikart G (2010) Genomics and the future of conservation genetics. Nat Rev Genet 11:697–710
Anderson CD, Ricks NJ, Farley KM et al (2016) Identification and characterization of microsatellite markers in Penstemon scariosus (Plantaginaceae). Appl Plant Sci 4:1–5
Bashalkhanov S, Pandey M, Rajora OP (2009) A simple method for estimating genetic diversity in large populations from finite sample sizes. BMC Genet 10:84
Brook BW, Tonkyn DW, O’Grady JJ, Frankham R (2002) Contribution of inbreeding to extinction risk in threatened species. Conserv Ecol 6(1):16
Caddah MK, Campos T, Zucchi MI, de Souza AP, Bittrich V, do Amaral MDE (2013) Species boundaries inferred from microsatellite markers in the Kielmeyera coriacea complex (Calophyllaceae) and evidence of asymmetric hybridization. Plant Syst Evol 299:731–741
Cahill AE, Levinton JS (2016) Genetic differentiation and reduced genetic diversity at the northern range edge of two species with different dispersal modes. Mol Ecol 25:515–526
Campbell D, Duchesne P, Bernatchez L (2003) AFLP utility for population assignment studies: analytical investigation and empirical comparison with microsatellites. Mol Ecol 12:1979–1991
Castellanos-Morales G, Gámez N, Castillo-Gámez RA, Eguiarte LE (2016) Peripatric speciation of an endemic species driven by Pleistocene climate change: the case of the Mexican prairie dog (Cynomys mexicanus). Mol Phylogenet Evol 94:171–181
Cavalli-Sforza LL, Edwards AWF (1967) Phylogenetic analysis models and estimation procedures. Am J Hum Genet 19:233–257
Cavers S, Degen B, Caron H, Lemes MR, Margis R, Salgueiro F, Lowe AJ (2005) Optimal sampling strategy for estimation of spatial genetic structure in tree populations. Heredity 95:281–289
Ceballos G, Ehrlich PR, Barnosky AD et al (2015) Accelerated modern human-induced species losses: entering the sixth mass extinction. Sci Adv 1:e1400253
Chybicki IJ, Oleksa A, Burczyk J (2011) Increased inbreeding and strong kinship structure in Taxus baccata estimated from both AFLP and SSR data. Heredity 107:589–600
Costa R, Pereira G, Garrido I, Tavares-de-Sousa M, Espinosa F (2016) Comparison of RAPD, ISSR, and AFLP molecular markers to reveal and classify orchardgrass (Dactylis glomerata L.) germplasm variations. PLoS ONE 11(4):e0152972
Crawford NG, Glenn T, Faircloth B et al (2010) SMOGD: software for the measurement of genetic diversity. Mol Ecol Resour 10:556–557
de Siqueira MF, Durigan G, de Marco Júnior P, Peterson AT (2009) Something from nothing: using landscape similarity and ecological niche modeling to find rare plant species. J Nat Conserv 17:25–32
De Barba M, Miquel C, Lobréaux S, Quenette PY, Swenson JE, Taberlet P (2017) High-throughput microsatellite genotyping in ecology: Improved accuracy, efficiency, standardization and success with low-quantity and degraded DNA. Mol Ecol Resour 17:492–507
Dockter RB, Elzinga DB, Geary B et al (2013) Developing molecular tools and insights into the Penstemon genome using genomic reduction and next-generation sequencing. BMC Genet 14:66
Duminil J, Fineschi S, Hampe A, Jordano P, Salvini D, Vendramin GG, Petit RJ (2007) Can population genetic structure be predicted from life-history traits. Am Nat 169:662–672
Earl DA, vonHoldt BM (2012) STRUCTURE HARVESTER: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv Genet Resour 4:359–361
El-Esawi MA, Germaine K, Bourke P, Malone R (2016) AFLP analysis of genetic diversity and phylogenetic relationships of Brassica oleracea in Ireland. C R Biol 339:163–170
Ellegren H (2004) Microsatellites: simple sequences with complex evolution. Nat Rev Genet 5:435–445
Ellstrand NC (1992) Gene flow by pollen: implications for plant conservation genetics. Oikos 1:77–86
England JL (1982) A new species of Penstemon (Scrophulariaceae) from the Uinta Basin of Utah and Colorado. Gt Basin Nat 42:367–368
Evanno G, Regnaut S, Goudet J (2005) Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol 14:2611–2620
Excoffier L, Lischer H (2010) Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Mol Ecol Resour 10:564–567
Fajardo D, Morales J, Zhu H et al (2013) Discrimination of American cranberry cultivars and assessment of clonal heterogeneity using microsatellite markers. Plant Mol Biol Rep 31:264–271
Falush D, Stephens M, Pritchard JK (2003) Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics 164:1567–1587
Felsenstein J (2005) PHYLIP (phylogeny inference package) version 3.6. Department of Genome Sciences, University of Washington, Seattle
Frankham R, Ballou J, Briscoe D (2010) Introduction to conservation genetics, 2nd edn. Cambridge University Press, New York
Galpern P, Peres-Neto PR, Polfus J, Manseau M (2014) MEMGENE: spatial pattern detection in genetic distance data. Methods Ecol Evol 5:1116–1120
Garcia AAF, Benchimol LL, Barbosa AMM et al (2004) Comparison of RAPD, RFLP, AFLP and SSR markers for diversity studies in tropical maize inbred lines. Genet Mol Biol 27:579–588
Gaudeul M, Till-Bottraud I, Barjon F, Manel S (2004) Genetic diversity and differentiation in Eryngium alpinum L.(Apiaceae): comparison of AFLP and microsatellite markers. Heredity 92:508–518
Goodrich S, Neese E (1986) Uinta Basin flora. U.S. Department of Agriculture, Forest Service, Intermountain Region, Ogden
Guisan A, Broennimann O, Engler R, Vust M, Yoccoz NG, Lehmann A, Zimmermann NE (2006) Using niche-based models to improve the sampling of rare species. Conserv Biol 20:501–511
Guo SW, Thompson EA (1992) Performing the exact test of Hardy-Weinberg proportion for multiple alleles. Biometric 48:361–372
Hale ML, Burg TM, Steeves TE (2012) Sampling for microsatellite-based population genetic studies: 25 to 30 individuals per population is enough to accurately estimate allele frequencies. PLoS ONE 7(9):e45170
Hamrick JL, Godt MW (1996) Effects of life history traits on genetic diversity in plant species. Phil Trans R Soc Lond 35:1291–1298
Helm A, Oja T, Saar L et al (2009) Human influence lowers plant genetic diversity in communities with extinction debt. J Ecol 97:1329–1336
Hilfiker K, Gugerli F, Schu J, Rotach P (2004) Low RAPD variation and female-biased sex ratio indicate genetic drift in small populations of the dioecious conifer Taxus baccata in Switzerland. Conserv Genet 5:357–365
Holmgren NH (1984) Penstemon. In: Cronquist A, Holmgren AH, Holmgren NH et al (eds) Intermountain flora: vascular plants of the intermountain West, U.S.A. asteridae except asteraceae, vol 4. New York Botanical Garden, Bronx, pp 370–457
Jakobsson M, Rosenberg NA (2007) CLUMPP: a cluster matching and permutation program for dealing with label switching and multimodality in analysis of population structure. Bioinformatics 23:1801–1806
Jarne P, Lagoda PJL (1996) Microsatellites, from molecules to populations and back. TREE 11:424–429
Jost L (2008) GST and its relatives do not measure differentiation. Mol Ecol 17:4015–4026
Jump AS, Peñuelas J (2007) Extensive spatial genetic structure revealed by AFLP but not SSR molecular markers in the wind-pollinated tree, Fagus sylvatica. Mol Ecol 16:925–936
Kaiser SA, Taylor SA, Chen N et al (2017) A comparative assessment of SNP and microsatellite markers for assigning parentage in a socially monogamous bird. Mol Ecol Resour 17:183–193
Kalia RK, Rai MK, Kalia S et al (2011) Microsatellite markers: an overview of the recent progress in plants. Euphytica 177:309–334
Kalinowski S (2005) Do polymorphic loci require large sample sizes to estimate genetic distances? Heredity 94:33–36
Kramer AT, Fant JB (2007) Isolation and characterization of microsatellite loci in Penstemon rostriflorus (Plantaginaceae) and cross-species amplification. Mol Ecol Notes 7:998–1001
Langella O (2007) Populations 1.2.30: population genetic software (individuals or population distances, phylogenetic trees). http://bioinformatics.org/;tryphon/populations/. Accessed 30 Jan 2017
Lee J (2016) Kennecott laying off 200 workers. DeseretNews.com, https://www.deseretnews.com/article/865649211/Kenn. Accessed 31 Oct 2017
Lewinsohn JS, Tepedino VJ (2007) Breeding biology and flower visitors of the rare White River penstemon, Penstemon scariosus var. albifluvis (Scrophulariaceae). West N Am Nat 67:232–237
Lomba A, Pellissier L, Randin C, Vicente J, Moreira F, Honrado J, Guisan A (2010) Overcoming the rare species modeling paradox: a novel hierarchical framework applied to an Iberian endemic plant. Biol Conserv 143:2647–2657
Lowe AJ, Boshier D, Ward M et al (2005) Genetic resource impacts of habitat loss and degradation; reconciling empirical evidence and predicted theory for neotropical trees. Heredity 95:255–273
Lynch M, Conery J, Burger R (1995) Mutation accumulation and the extinction of small populations. Am Nat 146:489–518
Maguire TL, Peakall R, Saenger P (2002) Comparative analysis of genetic diversity in the mangrove species Avicennia marina (Forsk.) Vierh. (Avicenniaceae) detected by AFLPs and SSRs. Theor Appl Genet 104:388–398
Meudt HM, Clarke AC (2007) Almost forgotten or latest practice? AFLP applications, analyses and advances. Trends Plant Sci 12:106–117
Michalski SG, Durka W (2012) Assessment of provenance delineation by genetic differentiation patterns and estimates of gene flow in the common grassland plant Geranium pratense. Conserv Genet 13:581–592
Miggiano E, De Innocentiis S, Ungaro A et al (2005) AFLP and microsatellites as genetic tags to identify cultured gilthead seabream escapees: data from a simulated floating cage breaking event. Aquacult Int 13:137–146
Mijangos JL, Pacioni C, Spencer PBS, Craig MD (2015) Contribution of genetics to ecological restoration. Mol Ecol 24:22–37
Morgante M, Olivieri AM (1993) PCR-amplified microsatellites as markers in plant genetics. Plant J 3:175–182
Neese EC (1986) New taxa and nomenclatural changes in Utah Penstemon (Scrophulariaceae). Gt Basin Nat 46:459–460
Newman D, Pilson D (1997) Increased probability of extinction due to decreased genetic effective population size: experimental populations of Clarkia pulchella. Evolution 51:354–362
Nold R (1999) Penstemons. Timber Press, Portland
Nybom H, Bartish IV (2000) Effects of life history traits and sampling strategies on genetic diversity estimates obtained with RAPD markers in plants. Perspect Plant Ecol Evol Syst 3:93–114
Oleksa A, Chybicki IJ, Gawron R (2013) Isolation by distance in saproxylic beetles may increase with niche specialization. J Insect Conserv 17:219–233
Ouborg NJ, Vergeer P, Mix C (2006) The rough edges of the conservation genetics paradigm for plants. J Ecol 94:1233–1248
Page RD (1996) TreeView: an application to display phylogenetic trees on personal computers. Comput Appl Biosci 12:357–358
Peakall R, Smouse PE (2012) GenAlEx 6.5: genetic analysis in Excel. Population genetic software for teaching and research—an update. Bioinformatics 28:2537–2539
Pennell F (1920) Scrophulariaceae of the central Rocky Mountain States, vol 20. United States National Herbarium, Washington, D.C., 313–381
Pimm SL, Raven P (2000) Extinction by numbers. Nature 403:843–845
Powell W, Morgante M, Andre C et al (1996) The comparison of RFLP, RAPD, AFLP and SSR (microsatellite) markers for germplasm analysis. Mol Breed 2:225–238
Pritchard JK, Seielstad MT, Perez-lezaun A, Feldman MW (1999) Population growth of human Y chromosomes: a study of Y chromosome microsatellites. Mol Biol Evol 16:1791–1798
Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155:945–959
Puechmaille SJ (2016) The program structure does not reliably recover the correct population structure when sampling is uneven: subsampling and new estimators alleviate the problem. Mol Ecol Resour 16:608–627
Putman AI, Carbone I (2014) Challenges in analysis and interpretation of microsatellite data for population genetic studies. Ecol Evol 22:4399–4428
Riek J, De CK, De Smulders MJM, Nybom H (2013) Molecular phylogenetics and evolution AFLP-based population structure analysis as a means to validate the complex taxonomy of dogroses (Rosa section Caninae). Mol Phylogenet Evol 67:547–559
Rocky Mtn. Wild, et al. v. W (2016) 15-cv-0615-WJM. U.S. District Court, D. Colorado. Order vacating administrative action and requiring meet-and-confer between the parties
Rodríguez-Peña RA, Jestrow B, Cinea W et al (2014a) Conservation and genetics of two critically endangered hispaniolan palms: genetic erosion of Pseudophoenix lediniana in contrast to P. ekmanii. Plant Syst Evol 300:2019–2027
Rodríguez-Peña RA, Jestrow B, Meerow AW et al (2014b) Genetic diversity and differentiation of Pseudophoenix (Arecaceae) in Hispaniola. Bot J Linn Soc 176:469–485
Rosenberg NA (2004) DISTRUCT: a program for the graphical display of population structure. Mol Ecol Notes 4:137–138
Rousset F (2008) GENEPOP ‘007: a complete re-implementation of the GENEPOP software for Windows and Linux. Mol Ecol Resour 8:103–106
Saccheri I, Kuussaari M, Kankare M et al (1998) Inbreeding and extinction in a butterfly metapopulation. Nature 392:491–494
Sinclair EA, Hobbs RJ (2009) Sample size effects on estimates of population genetic structure: implications for ecological restoration. Restor Ecol 17:837–844
Sønstebø JH, Borgstrøm R, Heun M (2007) A comparison of AFLPs and microsatellites to identify the population structure of brown trout (Salmo trutta L.) populations from Hardangervidda, Norway. Mol Ecol 16:1427–1438
Spielman D, Brook BW, Frankham R (2004) Most species are not driven to extinction before genetic factors impact them. Proc Natl Acad Sci 101:15261–15264
Sunnucks P (2000) Efficient genetic markers for population biology. TREE 15:199–203
Todd JJ, Vodkin LO (1996) Duplications that suppress and deletions that restore expression from a chalcone synthase multigene family. Plant Cell 8:687–699
U.S. Dept. of Interior - Fish and Wildlife Service (USFWS) (2013) Endangered and threatened wildlife and plants; threatened species status for Graham’s beardtongue (Penstemon grahamii) and White River beardtongue (Penstemon scariosus var. albifluvis); Proposed rule. Fed Regist 78:47590–47611
U.S. Dept. of Interior - Fish and Wildlife Service (USFWS) (2014) Endangered and threatened wildlife and plants; withdrawal of the proposed rules to list graham’s beardtongue (Penstemon grahamii) and White River beardtongue (Penstemon scariosus var. albifluvis) and designate critical habitat; Proposed rule. Fed Regist 79:46042–46087
U.S. Fish and Wildlife Service (FWS) (2017) Species listed in each state based on published historic range and population data. Environ Conserv Online Syst. https://ecos.fws.gov/ecp0/reports/species-listed-b. Accessed 1 Nov 2017
Van Oosterhout C, Hutchinson WF, Wills DPM, Shipley P (2004) MICRO-CHECKER: software for identifying and correcting genotyping errors in microsatellite data. Mol Ecol Resour 4:535–538
Vekemans X, Beauwens T, Lemaire M, Roldan-Ruiz I (2002) Data from amplified fragment length polymorphism (AFLP) markers show indication of size homoplasy and of a relationship between degree of homoplasy and fragment size. Mol Ecol 11:139–151
Von Kohn C, Conrad K, Kramer M, Pooler M (2018) Genetic diversity of Magnolia ashei characterized by SSR markers. Conserv Genet 19:923–936
Vos P, Hogers R, Bleeker M et al (1995) AFLP, a new technique for DNA fingerprinting. Nucleic Acids Res 23:4407–4414
Waycott M, Barnes G (2001) AFLP diversity within and between populations of the Caribbean seagrass Thalassia testudinum (Hydrocharitaceae). Mar Biol 139:1021–1028
Weir BS, Cockerham CC (1984) Estimating F-statistics for the analysis of population structure. Evolution 38:1358–1370
Wilcove DS, Rothstein D, Dubow J et al (1998) Quantifying threats to imperiled species in the United States. Bioscience 48:607–615
Wolfe AD, Randle CP, Datwyler SL et al (2006) Phylogeny, taxonomic affinities, and biogeography of Penstemon (Plantaginaceae) based on ITS and cpDNA sequence data. Am J Bot 93:1699–1713
Wolfe AD, McMullen-Sibul A, Tepedino VJ et al (2014) Conservation genetics and breeding system of Penstemon debilis (Plantaginaceae), a rare beardtongue endemic to oil shale talus in western Colorado, USA. J Syst Evol 52:598–611
Wolfe AD, Necamp T, Fassnacht S et al (2016) Population genetics of Penstemon albomarginatus (Plantaginaceae), a rare Mojave Desert species of conservation concern. Conserv Genet 17:1245–1255
Wright S (1951) The genetical structure of populations. Ann Eugen 15:323–354
Young A, Boyle T, Brown T (1996) The population genetic consequences of habitat fragmentation for plants. TREE 11:413–418
Acknowledgements
This research was funded by NFS DEB 1455399, and BLM Utah Grant # L14AC00346. The authors thank Jessica Brunson, Vernal Field Office, Bureau of Land Management, and Hope Hornbeck, SWCA Environmental Consultant, for assistance in collecting tissue samples from the field, and Paul Blischak, Benjamin Stone, and members of the Wolfe Lab for helpful comments on the manuscript. We also thank two anonymous referees for their comments on the manuscript.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Rodríguez-Peña, R.A., Johnson, R.L., Johnson, L.A. et al. Investigating the genetic diversity and differentiation patterns in the Penstemon scariosus species complex under different sample sizes using AFLPs and SSRs. Conserv Genet 19, 1335–1348 (2018). https://doi.org/10.1007/s10592-018-1103-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10592-018-1103-6