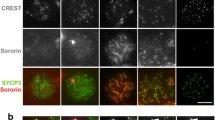
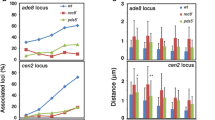
Abstract
Holocentric chromosomes occur in a number of independent eukaryotic lineages, and they form holokinetic kinetochores along the entire poleward chromatid surfaces. Due to this alternative chromosome structure, Luzula elegans sister chromatids segregate already in anaphase I followed by the segregation of the homologues in anaphase II. However, not yet known is the localization and dynamics of cohesin and the structure of the synaptonemal complex (SC) during meiosis. We show here that the α-kleisin subunit of cohesin localizes at the centromeres of both mitotic and meiotic metaphase chromosomes and that it, thus, may contribute to assemble the centromere in L. elegans. This localization and the formation of a tripartite SC structure indicate that the prophase I behaviour of L. elegans is similar as in monocentric species.




Similar content being viewed by others
Abbreviations
- dNTP:
-
Deoxynucleoside triphosphates
- PCR:
-
Polymerase chain reaction
- DNA:
-
Deoxyribonucleic acid
- SIM:
-
Structured illumination microscopy
- SMC:
-
Structural maintenance of chromosome
- SC:
-
Synaptonemal complex
- RACE:
-
Rapid amplification of cDNA ends
References
Albertson DG, Thomson JN (1993) Segregation of holocentric chromosomes at meiosis in the nematode, Caenorhabditis elegans. Chromosom Res 1:15–26
Anderson DE, Losada A, Erickson HP et al (2002) Condensin and cohesin display different arm conformations with characteristic hinge angles. J Cell Biol 156:419–424
Bhatt AM, Lister C, Page T et al (1999) The DIF1 gene of Arabidopsis is required for meiotic chromosome segregation and belongs to the REC8/RAD21 cohesin gene family. Plant J 19:463–472
Birkenbihl RP, Subramani S (1995) The rad21 gene product of Schizosaccharomyces pombe is a nuclear, cell cycle-regulated phosphoprotein. J Biol Chem 270:7703–7711
Cabral G, Marques A, Schubert V et al (2014) Chiasmatic and achiasmatic inverted meiosis of plants with holocentric chromosomes. Nat Commun 5:5070
Cai X, Dong F, Edelmann RE, Makaroff CA (2003) The Arabidopsis SYN1 cohesin protein is required for sister chromatid arm cohesion and homologous chromosome pairing. J Cell Sci 116:2999–3007
Calvente A, Barbero JL (2012) Cohesins and cohesin-regulators in meiosis. INTECH Open Access Publisher
Calvente A, Viera A, Parra MT et al (2013) Dynamics of cohesin subunits in grasshopper meiotic divisions. Chromosoma 122:77–91
da Costa-Nunes JA, Bhatt AM, O'Shea S et al (2006) Characterization of the three Arabidopsis thaliana RAD21 cohesins reveals differential responses to ionizing radiation. J Exp Bot 57:971–983
de Carvalho CE, Zaaijer S, Smolikov S et al (2008) LAB-1 antagonizes the Aurora B kinase in C. elegans. Genes Dev 22:2869–2885
Dong F, Cai X, Makaroff C (2001) Cloning and characterization of two Arabidopsis genes that belong to the RAD21/REC8 family of chromosome cohesin proteins. Gene 271:99–108
Eijpe M, Offenberg H, Jessberger R et al (2003) Meiotic cohesin REC8 marks the axial elements of rat synaptonemal complexes before cohesins SMC1β and SMC3. J Cell Biol 160:657–670
Garcia-Cruz R, Brieno MA, Roig I, et al (2010) Dynamics of cohesin proteins REC8, STAG3, SMC1β and SMC3 are consistent with a role in sister chromatid cohesion during meiosis in human oocytes. Human Reproduction: deq180.
Gerhard DS, Wagner L, Feingold EA et al (2004) The status, quality, and expansion of the NIH full-length cDNA project. Genome Res 14:2121–2127
Goldstein P (1987) Multiple synaptonemal complexes (polycomplexes): origin, structure and function. Cell Biol Int Rep 11:759–796
Golubovskaya IN, Hamant O, Timofejeva L et al (2006) Alleles of afd1 dissect REC8 functions during meiotic prophase I. J Cell Sci 119:3306–3315
Golubovskaya IN, Wang CJ, Timofejeva L, Cande WZ (2011) Maize meiotic mutants with improper or non-homologous synapsis due to problems in pairing or synaptonemal complex formation. J Exp Bot 62:1533–1544
Gong C, Li T, Li Q, Yan L, Wang T (2011) Rice OsRAD21-2 is expressed in actively dividing tissues and its ectopic expression in yeast results in aberrant cell division and growth. J Integr Plant Biol 53:14–24
Haering CH, Nasmyth K (2003) Building and breaking bridges between sister chromatids. BioEssays 25:1178–1191
Hartsuiker E, Vaessen E, Carr A, Kohli J (2001) Fission yeast Rad50 stimulates sister chromatid recombination and links cohesion with repair. EMBO J 20:6660–6671
Heckmann S, Schroeder-Reiter E, Kumke K, Ma L, Nagaki K, Murata M, Wanner G, Houben A (2011) Holocentric chromosomes of Luzula elegans are characterized by a longitudinal centromere groove, chromosome bending, and a terminal nucleolus organizer region. Cytogenet Genome Res 134:220–228
Heckmann S, Jankowska M, Schubert V, Kumke K, Ma W, Houben A (2014a) Alternative meiotic chromatid segregation in the holocentric plant Luzula elegans. Nat Commun 5:4979
Heckmann S, Schubert V, Houben A (2014b) Holocentric plant meiosis: first sisters, then homologues. Cell Cycle 13:3623–3624
Henderson KA, Keeney S (2005) Synaptonemal complex formation: where does it start? BioEssays 27:995–998
Herran Y, Gutierrez-Caballero C, Sanchez-Martin M et al (2011) The cohesin subunit RAD21L functions in meiotic synapsis and exhibits sexual dimorphism in fertility. EMBO J 30:3091–3105
Ishiguro K, Kim J, Fujiyama‐Nakamura S, Kato S, Watanabe Y (2011) A new meiosis‐specific cohesin complex implicated in the cohesin code for homologous pairing. EMBO Rep 12:267–275
Ishii T, Sunamura N, Matsumoto A et al (2015) Preferential recruitment of the maternal centromere-specific histone H3 (CENH3) in oat (Avena sativa L.) × pearl millet (Pennisetum glaucum L.) hybrid embryos. Chromosom Res 23:709–718
Jiang L, Xia M, Strittmatter LI, Makaroff CA (2007) The Arabidopsis cohesin protein SYN3 localizes to the nucleolus and is essential for gametogenesis. Plant J 50:1020–1034
Kaitna S, Pasierbek P, Jantsch M, Loidl J, Glotzer M (2002) The aurora B kinase AIR-2 regulates kinetochores during mitosis and is required for separation of homologous chromosomes during meiosis. Curr Biol 12:798–812
Klein F, Mahr P, Galova M, Buonomo SB, Michaelis C, Nairz K, Nasmyth K (1999) A central role for cohesins in sister chromatid cohesion, formation of axial elements, and recombination during yeast meiosis. Cell 98:91–103
Kudo NR, Wassmann K, Anger M et al (2006) Resolution of chiasmata in oocytes requires separase-mediated proteolysis. Cell 126:135–146
Kudo NR, Anger M, Peters AH et al (2009) Role of cleavage by separase of the Rec8 kleisin subunit of cohesin during mammalian meiosis I. J Cell Sci 122:2686–2698
Lee J, Hirano T (2011) RAD21L, a novel cohesin subunit implicated in linking homologous chromosomes in mammalian meiosis. J Cell Biol 192:263–276
Lee JY, Orr-Weaver TL (2001) The molecular basis of sister-chromatid cohesion. Annu Rev Cell Dev Biol 17:753–777
Lee J, Iwai T, Yokota T et al (2003) Temporally and spatially selective loss of Rec8 protein from meiotic chromosomes during mammalian meiosis. J Cell Sci 116:2781–2790
Llano E, Gómez R, Gutiérrez-Caballero C et al (2008) Shugoshin-2 is essential for the completion of meiosis but not for mitotic cell division in mice. Genes Dev 22:2400–2413
Losada A, Hirano M, Hirano T (2002) Cohesin release is required for sister chromatid resolution, but not for condensin-mediated compaction, at the onset of mitosis. Genes Dev 16:3004–3016
Marques A, Ribeiro T, Neumann P et al (2015) Holocentromeres in Rhynchospora are associated with genome-wide centromere-specific repeat arrays interspersed among euchromatin. Proc Natl Acad Sci 112:13633–13638
Michaelis C, Ciosk R, Nasmyth K (1997) Cohesins: chromosomal proteins that prevent premature separation of sister chromatids. Cell 91:35–45
Mito Y, Sugimoto A, Yamamoto M (2003) Distinct developmental function of two Caenorhabditis elegans homologs of the cohesin subunit Scc1/Rad21. Mol Biol Cell 14:2399–2409
Moraes IC, Lermontova I, Schubert I (2011) Recognition of A. thaliana centromeres by heterologous CENH3 requires high similarity to the endogenous protein. Plant Mol Biol 75:253–261
Nabeshima K, Villeneuve AM, Colaiácovo MP (2005) Crossing over is coupled to late meiotic prophase bivalent differentiation through asymmetric disassembly of the SC. J Cell Biol 168:683–689
Nagaki K, Kashihara K, Murata M (2005) Visualization of diffuse centromeres with centromere-specific histone H3 in the holocentric plant Luzula nivea. Plant Cell 17:1886–1893
Nasmyth K (2011) Cohesin: a catenase with separate entry and exit gates? Nat Cell Biol 13:1170–1177
Nasmyth K, Haering CH (2005) The structure and function of SMC and kleisin complexes. Annu Rev Biochem 74:595–648
Page SL, Hawley RS (2003) Chromosome choreography: the meiotic ballet. Science 301:785–789
Pasierbek P, Jantsch M, Melcher M, Schleiffer A, Schweizer D, Loidl J (2001) A Caenorhabditis elegans cohesion protein with functions in meiotic chromosome pairing and disjunction. Genes Dev 15:1349–1360
Qiao H, Lohmiller LD, Anderson LK (2011) Cohesin proteins load sequentially during prophase I in tomato primary microsporocytes. Chromosom Res 19:193–207
Sakuno T, Tada K, Watanabe Y (2009) Kinetochore geometry defined by cohesion within the centromere. Nature 458:852–858
Sanei M, Pickering R, Kumke K, Nasuda S, Houben A (2011) Loss of centromeric histone H3 (CENH3) from centromeres precedes uniparental chromosome elimination in interspecific barley hybrids. Proc Natl Acad Sci 108:E498–E505
Schägger H, Von Jagow G (1987) Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal Biochem 166:368–379
Schubert V (2009) SMC proteins and their multiple functions in higher plants. Cytogenet Genome Res 124:202–214
Schubert I, Dolezel J, Houben A, Scherthan H, Wanner G (1993) Refined examination of plant metaphase chromosome structure at different levels made feasible by new isolation methods. Chromosoma 102:96–101
Schubert V, Weissleder A, Ali H, Fuchs J, Lermontova I, Meister A, Schubert I (2009) Cohesin gene defects may impair sister chromatid alignment and genome stability in Arabidopsis thaliana. Chromosoma 118:591–605
Shao T, Tang D, Wang K et al (2011) OsREC8 is essential for chromatid cohesion and metaphase I monopolar orientation in rice meiosis. Plant Physiol 156:1386–1396
Smolikov S, Schild-Prufert K, Colaiácovo MP (2008) CRA-1 uncovers a double-strand break-dependent pathway promoting the assembly of central region proteins on chromosome axes during C. elegans meiosis. PLoS Genet 4, e1000088
Spurr AR (1969) A low-viscosity epoxy resin embedding medium for electron microscopy. J Ultrastruct Res 26:31–43
Sumara I, Vorlaufer E, Stukenberg PT et al (2002) The dissociation of cohesin from chromosomes in prophase is regulated by Polo-like kinase. Mol Cell 9:515–525
Suzuki G, Nishiuchi C, Tsuru A, Kako E, Li J, Yamamoto M, Mukai Y (2013) Cellular localization of mitotic RAD21 with repetitive amino acid motifs in Allium cepa. Gene 514:75–81
Sym M, Engebrecht J, Roeder GS (1993) ZIP1 is a synaptonemal complex protein required for meiotic chromosome synapsis. Cell 72:365–378
Tao J, Zhang L, Chong K, Wang T (2007) OsRAD21-3, an orthologue of yeast RAD21, is required for pollen development in Oryza sativa. Plant J 51:919–930
Valdeolmillos AM, Viera A, Page J et al (2007) Sequential loading of cohesin subunits during the first meiotic prophase of grasshoppers. PLoS Genet 3, e28
Waizenegger IC, Hauf S, Meinke A et al (2000) Two distinct pathways remove mammalian cohesin from chromosome arms in prophase and from centromeres in anaphase. Cell 103:399–410
Wang M, Tang D, Wang K, Shen Y, Qin B, Miao C, Li M, Cheng Z (2011) OsSGO1 maintains synaptonemal complex stabilization in addition to protecting centromeric cohesion during rice meiosis. Plant J 67:583–594
Weisshart K, Fuchs J, Schubert V (2016) Structured illumination microscopy (SIM) and photoactivated localization microscopy (PALM) to analyze the abundance and distribution of RNA polymerase II molecules on flow-sorted Arabidopsis nuclei. Bio-protocol 6, e1725
Westergaard M, von Wettstein D (1972) The synaptonemal complex. Annu Rev Genet 60:533–554
Xu H, Beasley M, Verschoor S et al (2004) A new role for the mitotic RAD21/SCC1 cohesin in meiotic chromosome cohesion and segregation in the mouse. EMBO Rep 5:378–384
Zetka MC, Kawasaki I, Strome S, Müller F (1999) Synapsis and chiasma formation in Caenorhabditis elegans require HIM-3, a meiotic chromosome core component that functions in chromosome segregation. Genes Dev 13:2258–2270
Zhang LR, Tao JY, Wang T (2004) Molecular characterization of OsRAD21-1, a rice homologue of yeast RAD21 essential for mitotic chromosome cohesion. J Exp Bot 55:1149–1152
Zhang L, Tao J, Wang S, Chong K, Wang T (2006) The rice OsRad21-4, an orthologue of yeast Rec8 protein, is required for efficient meiosis. Plant Mol Biol 60:533–554
Acknowledgments
We are grateful to all members of the Chromosome Structure & Function laboratory (IPK Gatersleben), Ingo Schubert (IPK) and E-Eva Tomaštíková (Centre of the Region Haná for Biotechnological and Agricultural, Research, Institute of Experimental Botany, Olomouc, Czech Republic) for fruitful discussions; to Karla Meier, Katrin Kumke, Oda Weiß, Isolde Tillack and Gresch Ulrike (IPK) for excellent technical assistance; to Anne Fiebig for sequence submission; and to Karin Lipfert (IPK) for help with artwork. This work was supported by the China CSC scholarship, the Deutsche Forschungsgemeinschaft (SPP 1384, HO 1779/17-1) and the IPK Gatersleben.
Author information
Authors and Affiliations
Corresponding author
Additional information
Responsible Editor: Jiming Jiang.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplemental Figure 1
CENH3 of L. elegans. (A) Gene structure model of LeCENH3, the positions of start/stop codons, and the in silico identified sequence. The obtained 5’ and 3’ RACE sequences and primer sites are indicated. (B) Alignment of CENH3 sequences from two Luzula species. The conserved domains of CENH3 are indicated by rectangle frames. (C) Phylogenetic analysis of CENH3 proteins from different species. (D) Western blot analysis using anti-LeCENH3, anti-histone H3 and anti-α-tubulin (as control) antibodies. The triangle indicates the band observed corresponding to the LeCENH3 protein. The total protein was extracted from L. elegans flower buds. (PDF 466 kb) (PDF 466 kb)
Supplemental Figure 2
The α-kleisins of L. elegans. (A) Phylogenetic analysis of α-kleisin-like proteins from different plant species. Reference IDs for the phylogenetic analysis of the α-kleisin sequences used in this study are available in Supplemental Table 2. (B) Protein structure model of four Luzula α-kleisin-like proteins based on in silico identification. The similarity of N-terminal conserved regions (red) among each other is indicated. (C) Gene structure model of Leα-kleisin-1 transcripts. Positions of start/stop codon and primer sites are indicated. (D) The purified recombinant Leα-kleisin protein was analyzed by Coomassie staining (blue gel on left) and Western blotting (black picture on right) with anti-6X His tag antibodies. The major band observed corresponds to the Leα-kleisin protein (triangle). (E) The purified recombinant Leα-kleisin protein was analyzed by Leα-kleisin recombinant antibody. The major band observed corresponds to the Leα-kleisin protein (triangle). (PDF 589 kb)
Supplemental Figure 3
The distribution of Leα-kleisin (red) and LeCENH3 (green) at pachytene of L. elegans was identified by SIM. The panels below show the regions of interest (rectangle) further magnified. (PDF 94 kb)
Supplemental Figure 4
The distribution of anti-OsSGO1 (red) along metaphase II chromosomes of L. elegans. (PDF 43 kb)
Supplemental Figure 5
The centromere localization of Leα-kleisin (red) in somatic metaphase chromosomes of V. faba was identified by SIM. (PDF 119 kb)
Supplemental Table 1
List of primer sequences for PCR, RT-PCR and FISH. (DOCX 18 kb)
Supplemental Table 2
List of sequence identifiers and description of α-kleisin sequences used for phylogenetic tree construction. (DOCX 17 kb)
Supplemental Table 3
The similarity of different Leα-kleisin protein sequences. (DOCX 17 kb)
Rights and permissions
About this article
Cite this article
Ma, W., Schubert, V., Martis, M.M. et al. The distribution of α-kleisin during meiosis in the holocentromeric plant Luzula elegans . Chromosome Res 24, 393–405 (2016). https://doi.org/10.1007/s10577-016-9529-5
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10577-016-9529-5