Abstract
Purpose
Breast cancer is one of the leading types of cancer diagnosed in women. Despite the improvements in chemotherapeutic cure strategies, drug resistance is still an obstacle leading to disease aggressiveness. The small non-coding RNA molecules, miRNAs, have been implicated recently to be involved as regulators of gene expression through the silencing of mRNA targets that contributed to several cellular processes related to cancer metastasis. Hence, the present study aimed to investigate the beneficial role and mechanism of miRNA-34a-based gene therapy as a novel approach for conquering drug resistance mediated by ATP-binding cassette (ABC) transporters in breast cancer cells, besides exploring the associated invasive behaviors.
Material and Methods
Bioinformatics tools were used to predict miRNA ABC transporter targets by tracking the ABC transporter pathway. After the establishment of drug-resistant breast cancer MCF-7 and MDA-MB-231 sublines, cells were transfected with the mimic or inhibitor of miRNA-34a-5p. The quantitative expression of genes involved in drug resistance was performed by QRT-PCR, and the exact ABC transporter target specification interaction was confirmed by dual-luciferase reporter assay. Furthermore, flow cytometric analysis was utilized to determine the ability of miRNA-34a-treated cells against doxorubicin uptake and accumulation in cell cycle phases. The spreading capability was examined by colony formation, migration, and wound healing assays. The apoptotic activity was estimated as well.
Results
Our findings firstly discovered the mechanism of miRNA-34a-5p restoration as an anti-drug-resistant molecule that highly significantly attenuates the expression of ABCC1 via the direct targeting of its 3′- untranslated regions in resistant breast cancer cell lines, with a significant increase of doxorubicin influx by MDA-MB-231/Dox-resistant cells. Additionally, the current data validated a significant reduction of metastatic potentials upon miRNA-34a-5p upregulation in both types of breast cancer-resistant cells.
Conclusion
The ectopic expression of miRNA-34a ameliorates the acquired drug resistance and the migration properties that may eventually lead to improved clinical strategies and outcomes for breast cancer patients. Additionally, miRNA-34a could be monitored as a diagnostic/prognostic biomarker for resistant conditions.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Breast cancer (BC) is the most prevalent cancer type among women and the mainly common malignant tumor in the world among all cancers. BC affects females at any age after puberty in every country around the globe, with rising incidence in later life. In 2020, there were 2.3 million women diagnosed with BC and 685,000 deaths worldwide [1]. According to the glance statistics of the National Cancer Institute, there were 297,790 estimated new BC cases in the year 2023, representing 15.2% of all new cancer cases [2]. Despite the availability of many early detection and treatment options, as well as advances in understanding the molecular mechanisms of BC biology, the disease continues to be fatal and metastatic as a result of rapid development of resistance to cytotoxic medicines and alterations in many gene expressions [3,4,5]. Consequently as identifying novel effective treatment methods is currently urgent in the field of breast cancer research to improve the clinical management of cancer, adjustment of proliferation, apoptosis, angiogenesis, metastasis, and drug resistance mediatory factors by gene expression regulators has been used as a competence for treatment [6,7,8].
MicroRNAs (miRNAs) are very important gene expression regulators that have a vital role in controlling many cellular biological processes such as cell growth, proliferation, invasion, differentiation, adhesion, cell death, drug sensitivity/resistance, and cellular metabolic pathways. They are highly conserved naturally occurring small non-coding RNA molecules with a length of about 21–24 nucleotides which can bind partially to their complementary bases in the 3′- untranslated regions (3′UTR) of protein-coding mRNAs, resulting in mRNA translation inhibition or destabilization, leading to gene silencing at the post-transcriptional level [6, 9,10,11].
MiRNA-34a is one of the miRNA-34 family members that contains three processed miRNAs and are encoded by two different genes. The locus encoding miR-34a is located on chromosome 1p36, while miR-34b and miR-34c share primary transcription from chromosome 11q23. MiR-34a has attracted the greatest attention in the cancer research community because it functions as a tumor suppressor gene; it is also the first miRNA to be shown to be directly regulated by the tumor suppressor p53. MiR-34a is highly expressed in normal cells as compared to carcinomas in which its expression is often downregulated [10, 12, 13]. Because of its critical role in several oncogenic pathways of cancer types, including apoptosis and cell proliferation, miRNA-34a has recently motivated the interest of numerous researchers. Moreover, miRNA-34a has been established to be associated with drug resistance in BC [14,15,16].
Drug resistance, whether endogenous or acquired, is an important obstacle to cancer treatment success, resulting in an inefficient chemotherapeutic strategy and, ultimately, mortality. There are a number of mechanisms that mediate drug resistance in cancer cells, but the most widely studied one is drug efflux through the overexpressed cell membrane ATP-binding cassette (ABC) transporters that determine the rate of anticancer drugs pumping outside the cells. These drug pumping regulators include P-gp (P-glycoprotein, ABCB1), multidrug resistance-associated protein 1 (ABCC1), ABCC5, ABCC10, ABCF2, and breast cancer resistance protein, ABCG2 (BCRP) [16, 17].
Although several roles of miRNAs in cancer were recognized, the role played by miRNAs in drug resistance is still not fully elucidated [18, 19]. Therefore, the present work aimed to study the therapeutic anti-tumor effects of miRNA-34a restoration in established resistant MCF-7/tamoxifen and MDA-MD-231/doxorubicin cell lines. As well as investigating the mechanism of drug resistance reversal that may be promoted by miRNA-34a upregulation to facilitate the improvement of medicine efficacy and patient response to diverse treatments.
Material and methods
Bioinformatics analysis
As described by Kern et al. [20] and Yang et al. [21], the pathway prediction was carried out by miRWALK (http://mirwalk.umm.uni-heidelberg.de) for the ABC transporters investigation using gene-miRNA-pathway analysis tool, the predicted targets were further elucidated by Target scan and RNA22. The mature sequence of miRNA-34a-5p was downloaded from miRBase and the 3'UTR sequences of human ABC transporter genes were downloaded from the Ensembl genome browser.
Cell lines and cell culture
According to Rana et al. [22], wild MCF-7 (ATCC HTB-22) (human non-invasive luminal type breast cancer) and MDA-MD-231 (ATCC HTB-26) (aggressive human triple-negative breast cancer) human breast cancer cell lines were purchased from ATCC (American Type Culture Collection, VA, USA). Cells were cultured and propagated in 75 cm2 flasks in DMEM (Dulbecco’s Modified Eagle Medium; Lonza, Belgium); supplemented with 10% fetal bovine serum (FBS; Biochrom, Berlin), 1% penicillin–streptomycin (Lonza, Belgium), and 4 mM L glutamine (Lonza, Belgium) at 37 ℃ in 5% CO2 incubator. When the cell density reached approximately 80–100%, cells were split for further cultures.
Establishment of drug-resistant cells
Drug resistance was induced in human breast cancer cells as reported by Krisnamurti et al. [23]. Briefly, MCF-7 and MDA-MB-231 breast cancer cells were treated with tamoxifen and doxorubicin at doses of 1 and 0.18 μM, respectively, for 10 passages. The dramatic upregulation of ABCB1 and ABCG genes to near two or three folds, as well as the morphological changes, confirmed the resistance induction.
Transfection of cells with microRNA-34a
Hsa-miRNA-34a-5p mimic, inhibitor, and miscript miRNA negative control (NC) were purchased from Qiagen (USA). Both resistant cell lines MCF-7/ tamoxifen (MCF-7/Tx), and MDA-MB-231/Doxorubicin (MDA-MB-231/Dox) were treated with miRNA-34a-5p mimic, or inhibitor, or NC using Hiperfect transfection reagent (Qiagen, USA) according to the following protocol described by Yahya et al. [24]; before transfection, 1.5 × 105 cells were seeded/ well onto two 24-well plates in 500 µl of growth DMEM culture medium. On the second day, cells were transfected with 50 nM miRNA-34a-5p mimic, or inhibitor, or NC in serum-free media using 1 µl of the Hiperfect transfection reagent. After 24 h, the growth media were changed with fresh ones and the cells in the first plate were incubated at the proper experimental conditions for an additional 24 h. The cells in the second plate were subjected to miRNA extraction to verify successful transfection.
Qiagen miscript system for miRNA-34a determination
MiRNA-34a-5p was determined using the Qiagen miscript system (Qiagen, USA) as directed by the manufacturer. In brief, the miRNeasy kit was used to purify RNA-containing miRNAs, and the miscript RT-kit was used to generate cDNA from the purified RNA-containing miRNAs. In separate procedures, real-time PCR was used to detect mature miRNA utilizing miRNA-specific primers targeting hsa-miRNA-34a-5p. The cycling conditions were as follows: denaturation for 15 s at 94 °C, annealing for 30 s at 55 °C, and extension for 30 s at 70 °C. According to Yahya et al. [24], fluorescence data were obtained using a MiniOpticon Bio-Rad Real-Time Thermal Cycler (France).
Quantitative real-time gene expression analysis
This was performed using a SYBR® Green one-step RT-PCR kit (Qiagen, USA) after extraction of total RNAs from 1.5 × 105 MCF-7/Tx and MDA-MB-231/Dox cells. The resistant cells treated with miRNA-34a mimic or inhibitor or NC were lysed by QIAzol lysis reagent (Qiagen, USA) according to the manufacturer's instructions. The primer sequences of targeted genes are shown in Table 1. Each gene copy number was normalized to 100,000 copies of the β-actin gene. The RT and subsequent PCR cycling condition was as follow: 50 °C for 10 min, 95 °C for 5 min, 60 °C for 30 s, and then 95 °C for 15 s; the number of cycles was 40 cycles. Bio-Rad Miniopticon™ real-time PCR cycler was used for quantitative estimation [24,25,26].
Dual-luciferase reporter assay and the vector construction
Referring to Dickson et al. [27], oligonucleotides corresponding to the surrounding predicted target sequence of miRNA-34a-5p in 3' UTR of ABCB1, ABCC1, and ABCC5 were designed and cloned into the Dra1 and Xho1 sites of pmirGLO Dual-Luciferase miRNA Target Expression Vector (Promega, USA) which was linearized with the same restriction enzymes then ligated with ligase enzyme. Insertion was verified by PCR (primers and oligonucleotide sequences are listed in Table 2). MCF-7 cells (10,000 cells) were co-transfected with the vector of each construct and miScript negative control (NC treatment), or, mir-34a-5p mimic. After co-transfection for 48 h, the relative luciferase activity was measured using a dual-Glo Luciferase Assay System (Promega, USA). The Dual-Glo® Luciferase Assay System is designed to allow high-throughput analysis of mammalian cells containing genes for firefly and Renilla luciferases grown in 96-well plates. The firefly luciferase luminescence was divided over Renilla luciferase luminescence of each treatment, and then it was calculated as % of control treatment corresponding to its vector construct [21, 28].
Influx assay
Influx assay was accomplished as stated by Punia et al. [29], with some modifications. Briefly, 2 × 106 MDA-MB-231/Dox cells were transfected with miRNA-34a-5p mimic or NC for 48 h. Then, cells were incubated with 10 µM doxorubicin in phosphate-buffered saline (PBS) for 2 h. Next, cells were harvested, washed, and re-suspended in PBS. Doxorubicin uptake by the cells was examined using a NovoCyte flowcytometry (Agilent, USA) and analyzed by NovoExpress 1.4.1. software. Control background cells were used as a background and its fluorescence measurements were subtracted from all treatment values.
Flow cytometric analysis of cell cycle distribution
The DNA content was used to examine cell cycle distribution using the propidium iodide (PI) staining method. Doxorubicin-resistant MDA-MB-231 cells were grown at a density of 1 × 106 cells/ well onto a six-well plate and then transfected with miscript miRNA negative control or miR-34a-5p mimics for 48 h. Cells were then washed with Dulbecco’s phosphate-buffered saline (DPBS), trypsinized using 0.05% trypsin–EDTA, fixed in 70% ethanol in DPBS, and stored at 4 °C overnight. Cells were incubated with 50 µg/ ml PI (Thermo Scientific) containing 8 µg/ml RNase A in the dark at 37 °C for 30 min [24, 26, 30]. Cells were then analyzed by NovoCyte flowcytometer (Agilent, USA). The percentage of cells in G1, S, G2/M and, sub-G1 phases of the cell cycle was calculated using NovoExpress 1.4.1. software.
Colony formation assay
Both MCF-7/Tx and MDA-MB-231/Dox Cells were cultured for 14 days in 6-well plates with a density of 2 × 103 cells/well after being transfected with miRNA-34a-5p mimics or NC. Colonies were then washed with DPBS, fixed with 1:7 acetic acid/methanol for 5 min at room temperature, washed again with DPBS, dyed with 0.5% crystal violet for 30 min, then washed with tap water and left to air dried [31]. After that, it was captured for counting using an inverted fluorescence microscope (Zeiss, Germany).
Transwell migration assay
After 48 h of miRNA-34a-5p mimic/ NC transfection, migration assay was performed using polycarbonate membranes with 8-μm pores (Thincert™, Greiner Bio-One, Germany) in 24-well plates. MCF-7/Tx and MDA-MB-231/Dox Cells were serum-starved by incubating cells in media containing 0.2% FBS and then kept at 37 °C in a 5% CO2 incubator for 24 h. At a density of 1 × 104 cells/ml in 100 μl of serum-free medium, cells were placed in the upper chamber of the transwell assembly while the lower chamber contained 650 μl of 5% FBS-growth media. After incubation at 37 °C and 5% CO2 for 24 h, the upper surface of the membrane was scraped gently to remove non-migrating cells and washed with PBS. The membrane was then fixed in 4% paraformaldehyde for 15 min and stained with crystal violet. The cells were then imaged in five fields for each membrane and counted using Image J software [32].
Wound healing assay
After miRNA-34a-5p mimic/ NC transfected cells (MCF-7/ Tx and MDA-MB-231/Dox) reached confluency, a 100 µl pipette tip was applied to create a homogeneous scratch wound on the monolayer. Cells were photographed 24 h after creating the scratch. The width of scratches was quantified under an inverted microscope (Zeiss, Germany) at three different positions (bottom, middle, and top), and the mean width was calculated [33].
Annexin assay for apoptosis and necrosis measurement
These were measured using The RealTime-Glo™ Annexin V Apoptosis and Necrosis Assay (Promega, USA) [34]. Briefly, 10 × 103 cells (MCF-7/ Tx and MDA-MB-231/ Dox)/ well were seeded onto a 96 opaque black plate. The next day cells were transfected with miRNA-34a-5p mimic or NC, and the detection reagent of the RealTime-Glo™ Annexin V Apoptosis and Necrosis Assay which contains Annexin V NanoBiT™ Substrate, CaCl2, Necrosis Detection Reagent, Annexin V-SmBiT and Annexin V-LgBiT reagents was dissolved in cell growth media containing 5% FBS. Six hours after transfection, RLU (Relative Light Unit) and RFU (Relative Fluorescent Unit) were measured using a plate reader (FluoStar, Optima BMG, Germany), after that, FLU (Florescent and luminescent unit) and RFU were recorded at different time intervals for the two consecutive days. A graph was plotted which plots RLU and RFU development against time.
Caspase 3/7 measurement
The Caspase-Glo® 3/7 Assay (Promega, USA) is a homogeneous, luminescent assay that measures caspase-3 and -7 activities. Briefly, 10,000 cells (MCF-7/ Tx and MDA-MB-231/ Dox) were treated with miRNA-34a-5p mimic or NC for 48 h. After that, a 100 µl of Caspase-Glo® 3/7 Reagent was added to each well, and luminescent was measured using FluoStar, Optima BMG plate reader (Germany) [34].
Statistical analysis
The results were presented as mean±SEM (standard error of the mean) after their analysis by GraphPad Prism version 5.0 (GraphPad Software Inc., San Diego, CA, USA). A Student’s t-test was used to compare individual data points among each group. A P value of < 0.05 was set as the criterion for statistical significance. The reproducibility of the data was confirmed as each experiment was performed in triplicate three independent times.
Results
Confirmation of miRNA-34a-5p transfection in drug-resistant human breast cancer cells
After transfection of miRNA-34a-5p mimic/inhibitor/NC, miRNA-34a-5p levels were determined in MCF-7/Tx and MDA-MB-231/Dox cells to verify the success of transfection. As evident in Fig. 1A, miRNA-34a-5p expression levels were dramatically decreased in miRNA-34a inhibitor-treated MCF-7/TX and MDA-MB-231/Dox cells as compared to the negative control (NC) treatment. While miRNA-34a-5p expression levels were obviously up-regulated in miRNA-34a mimics-treated MCF-7/TX and MDA-MB-231/Dox cells as compared to the NC treatment (Fig. 1B).
Verification of the predicted drug resistance-related target genes by Dual-Luciferase reporter assay and QRT-PCR
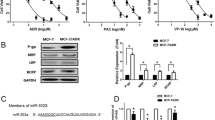
MiRWALK (http://mirwalk.umm.uni-heidelberg.de) analysis using gene-miRNA-pathway analysis tool, Target scan, and RNA22 bioinformatics tools have confirmed that ABCB1, ABCC1, and ABCC5 are predicted targets for miRNA-34a (Fig. 2A). To verify this prediction experimentally, in addition to real-time PCR, dual–luciferase reporter assay was conducted. Insertion of the ABCC1 binding site into the vector and co-transfection with miRNA-34a-5p mimics significantly reduced the luciferase activity, whereas insertion of ABCB1 and ABCC5 binding sites into vector and co-transfection with miRNA-34a-5p mimics produced a non-significant decrease in the luciferase activity (Fig. 2B).
MiRNA-34a-5p targets ABCB1, ABCC1, and ABCC5; A Schematic representation of bioinformatics analysis of predicted consequential pairing of ABCB1, ABCC1, and ABCC5 3` UTR targeting sequence and miRNA-34a-5p seed region; B Dual-Luciferase reporter assay indicated that MiRNA-34a can bind to 3' UTR of ABCC1; C ABC transporters gene expression levels in miRNA-34a-5p inhibitor MCF-7/Tx-treated cells; D ABC transporters gene expression levels in miRNA-34a-5p inhibitor MDA-MB-231/Dox-treated cells; E ABCB1 transporter gene expression levels in miRNA-34a-5p mimic and NC-treated MCF-7/Tx and MDA-MB-231/Dox cells; F ABCC1 transporter gene expression levels in miRNA-34a-5p mimic and NC transfected MCF-7/Tx and MDA-MB-231/Dox cells; G ABCC5 transporter gene expression levels in miRNA-34a-5p mimic and NC transfected MCF-7/Tx and MDA-MB-231/Dox cells. P < 0.05 (*), *** < 0.001
As shown in Fig. 2C, miRNA-34a-5p inhibition resulted in a significant upregulation of ABCB1, ABCC1, and ABCF2 gene expression levels in MCF-7/Tx cells; while in MDA-MB-231/ Dox cells (Fig. 2D), the expression level of ABCF2 gene was recorded to be significantly increased as compared to the other genes expression levels.
On the other hand, treatment of MCF-7/Tx and MDA-MB-231/Dox cells with miR-34a-5p mimics revealed a highly significant (P < 0.001) decrease in levels of ABCC1 gene as compared to NC-treated cells (Fig. 2F). Meanwhile, it produced a significant downregulation of ABCC5 gene expression level in MDA-MB-231/Dox cells (Fig. 2G). However, the changes were insignificant in the ABCB1 gene as compared to the NC treatment (Fig. 2E).
Upregulation of miRNA-34a-5p increases doxorubicin uptake by resistant breast cancer cells
Doxorubicin influx was detected to be significantly retained in MDA-MB-231/Dox cells treated with miRNA-34a-5p mimics by flow cytometry, in a comparison with NC, as displayed in Fig. 3A and B.
Doxorubicin influx measured by flow cytometry in MDA-MB-231/Dox cells. A a. Flow cytometric analysis of control background cells, b. Miscript negative control (NC)-treated cells, c. MiRNA-34a-5p mimic-treated cells. B Doxorubicin mean florescent in cells incubated with miRNA-34a-5p mimic for 48 h. (*P < 0.05)
The effect of miRNA-34a-5p overexpression on cell cycle progression
As mir-34a-5p downregulation has been observed in many different tumors including breast cancer [35, 36], we increased its concentration within doxorubicin-resistant breast MDA-MB-231 cancer cells by transfection of mir-34a-5p mimics. The results of flow cytometry reported the insignificant effect of this microRNA on cell cycle distribution. The observations noticed a slight increase in the mean percentage of cells in G0/G1 (37 ± 1.07%), and G2/M (23.84 ± 1.8%) phases as compared to the corresponding controls (36 ± 0.82% and 22 ± 2.7%, respectively). At the same time, our results showed a decrease in the accumulation of cells in the sub-G1 phase after 48 h transfection of mir-34a-5p mimics, in a comparison to control (Fig. 4A, B and Table 3).
Cell cycle distribution analysis by flow cytometry. A Representative data of flow cytometric analysis of MDA-MB231/Dox cells treated with miRNA-34a-5p mimic/NC for 48 h. The analysis was carried out by NovoCyte flow cytometer. B A column chart of percentage of accumulated cells in each cell cycle phase as a response to miRNA-34a-5p upregulation in MDA-MB231/Dox cells
Malignant biological behavior of miRNA-34a-5p related to progression and invasion in human-resistant breast cancer cells
To investigate the anti-proliferative therapeutic function of miRNA-34a-5p restoration in drug-resistant breast cancer cell lines, MCF-7/Tx and MDA-MB-231, colony formation, transwell migration, and wound healing assays were performed. Results of the colony formation (Fig. 5), the migration ability (Fig. 6 and Table 4), and wound healing (Fig. 7) showed a significant attenuation of metastatic potential and cellular growth suppression upon miRNA-34a-5p upregulation in both types of breast cancer-resistant cells.
Migration inhibition of MCF-7/Tx and MDA-MB-231/Dox cells in response to exogenous manipulation of miRNA-34a-5p expression. A, and B Representative images of migrated cells after transfection of MCF-7/Tx, and MDA-MB-231/Dox cells with miR-34-a-5p mimic for 48 h using transwell migration assay, respectively, and C The effect of miRNA-34a-5p inhibitor treatment on migration in MDA-MB-231/Dox cells
The apoptotic and necrotic activities of miRNA-34a-5p
In the current study, both annexin apoptosis assay and caspase-3/7 activity didn’t confirm a remarkable effect of miRNA-34a-5p restoration on apoptosis (Figs. 8 and 9). In annexin assay, miRNA-34a-5p mimic-treated cells (MCF-7/Tx and MDA-MB-231/Dox) produced time- and dose-dependent upregulation in luminescence which was concurrent with increased inflorescence which did augment neither apoptotic nor necrotic effects and suggested the involvement of alternative mechanisms of programmed cell death.
Annexin assay for determination of apoptosis and necrosis. A MCF-7/Tx cells transfected by miRNA-34a-5p mimic/NC, and B MDA-MB-231/ Dox cells treated with miscript miRNA-34a-5p negative control (NC), miRNA-34a-5p mimic. RLU Relative Light Unit, and RFU Relative Fluorescent Unit. Data were presented as mean ± SEM
Discussion
Breast cancer (BC) is the main cause of cancer mortality among women globally. According to statistics, BC accounts for around 23% of all cancer cases in women, and death due to breast carcinoma accounts for approximately 14% of all cancer-related mortalities in women, while the number of BC patients is increasing at a rate of more than 1.3 million per year. The poor response to chemotherapy due to the development of cellular drug resistance remains a key clinical obstacle to successful BC treatment [37, 38].
One of the most important mechanisms of drug resistance is drug elimination through the ATP-binding cassette (ABC) transporter family, which reduces intracellular drug accumulation by working as a pump rapidly extruding exogenous chemicals out of cells. The ABC family members including ABCB1, ABCC1, ABCC2, and ABCG2 were also expressed to overlap in substrate recognition, making those transmembrane transporters one of the leading causes of medication failure [19].
MiRNAs play an important role in targeting specific genes related to chemotherapeutic resistance, and multiple of them target several genes contributed to different attributed biochemical pathways. Various studies have shown that rebalancing miRNA expression in resistant cells might re-sensitize cancer cells to anti-tumor therapies, bolstering the potential of miRNAs as co-adjuvants in anticancer treatments. MiRNA-34a, the master regulator of tumor suppression, was discovered to be downregulated in a variety of malignancies, including BC [6, 32]. Furthermore, miRNA-34a-5p, derived from miRNA-34a, has been perceived to inhibit cell migration, invasion, and tumor development [14] and suggested a correlated mechanism between miRNA-34a and anti-apoptotic proteins [39]. The strategy of miRNA-34a replacement has been explored in clinical trials as the first challenge of miRNA application in cancer treatment [40]. Therefore, the current research paper firstly underlined the potential of miRNA-34a-5p upregulation as a drug resistance modulator in established resistant BC cell lines and studied the anticancer therapeutic effects of miRNA-34a-5p restoration in MCF-7/ tamoxifen and MBA-MB-231/doxorubicin-resistant BC cells. Interestingly, our findings indicated that the overexpression of miRNA-34a-5p increased the sensitivity of resistant cells in a mechanism that is dependent on direct targeting the drug transporter ABCC1 gene, as confirmed by dual-luciferase reporter assay, QRT-PCR, and influx assay. Moreover, the transfection of miRNA-34a mimic led to diminishing the metastatic and invasive properties of resistant BC cells, as observed by colony formation, transwell migration, and wound healing assays. However, our results suggested that there might be more active anti-proliferative mechanisms than apoptosis and cell cycle arrest.
An agreement with our data, He et al. [18] and Shao et al. [41] declared that miRNA-34a improves the sensitivity of cervical cancer cells toward oxaliplatin by targeting Murine Double Minute 4 (MDM4) expression and 5-fluorouracil by targeting lactate dehydrogenase A (LDHA), a glycolysis key enzyme, respectively. Moreover, overexpression of hsa-miRNA-34a-5p in gastric cancer SGC-7901/5-fluorouracil-resistant cells was found to decrease migration and invasiveness, as resulted by wound healing and transwell invasion assays, after chemotherapy. Likewise, hsa-miR-34a-5p enhanced the chemotherapy sensitivity of resistant gastric cancer cells by targeting SIRT1, ABCB1, and ABCC1 [42]. However, the current results showed a non–significant reduction in luciferase activity in the case of ABCB1 and ABCC5 3’UTR vector co-transfection with hsa-miR-34a-5p mimic. In addition, overexpression of hsa‑miRNA‑34a was detected to inhibit cell migration and invasion in bladder cancer cell lines 5637 and UMUC3 as detected by transwell assay, through matrix metalloproteinase-2 silencing [43]. Han et al. [33] confirmed that the treatment of T‑47D and MDA‑MB‑231 breast cancer cells with miRNA-34 mimic inhibited tumor cell proliferation, migration, and invasiveness as demonstrated by Colony formation, Wound healing, and transwell invasion assays. While upregulation of miRNA-34a in both these breast cancer cell lines increased the activity of caspase-3 via downregulation of stem cell-associated transcription factors, E2F1 and E2F3. MiRNA-34a mimic was determined to inhibit the proliferation and colony formation of SKOV3 and OVCA433 ovarian cancer cells and enhanced cisplatin sensitivity of cisplatin-resistant SKOV3 ovarian cells via direct suppressing of HDAC1 [44]. The ectopic expression of miRNA-34a in metastatic breast cancer cell-line BT-549 significantly inhibited cell migration and invasion by directly targeting epithelial to mesenchymal transition (EMT)-associated protein NOTCH1 [45].
More conformance oncology studies were reported in a research work conducted by Williams et al. [46] demonstrated that miRNA-34a mimic reversed cisplatin and vinorelbine resistance in malignant pleural mesothelioma cell lines and enhanced apoptosis. As well, upregulation of miRNA-34a resulted in attenuating drug resistance to 5-fluorouracil in colon cancer via downregulation of targets in the PI3K/AKT signaling pathway. Also, miRNA-34a restoration improved cisplatin sensitivity in bladder cancer cell lines via transcriptional suppression of SIRT1 and Cdk6. Besides, increasing miRNA-34b expression enhanced susceptibility to gemcitabine, doxorubicin, and methotrexate, as well as triggered apoptosis. This was most likely accomplished by miR-34b modulation of PAK1 and ABCB1, which are important regulators of the cell cycle, multidrug resistance (MDR), and apoptosis [46].
A study performed by Liu and co-workers [47] agreed with our observations. They detected that miRNA-34a mimic attenuated the paclitaxel-chemoresistance of PC-3PR prostate cancer cells through direct suppression of JAG1 and Notch1expressions. Overexpression of miRNA-34a reduced the formation of prostate cancer cell clones as well. While high levels of miRNA-34a promoted cellular apoptosis and G0/G1 arrest of PC-3PR cells [47]. A direct interaction of miRNA-34a with the anti-apoptotic BCL-2 gene was shown by luciferase assay in MCF-7, and MDA-MB-231 acquired docetaxel resistance [4]. Additionally, Liu and co-workers in 2012 suggested that the upregulation of miRNA-34a in prostate cancer cells was associated with the increased sensitivity of tumor cells to camptothecin and paclitaxel cytotoxic agents [38].
The mechanism by which miR-34a targets ATP-binding cassette (ABC) transporters could be explained by its direct targeting of the 3′UTR of the ornithine decarboxylase antizyme 2 (OAZ2), resulting in suppression of MDR-associated ABC transporters. This finding was reported earlier in colorectal adenocarcinoma where the ectopic expression of exogenous miRNA-34a re-sensitized MDR-human colorectal adenocarcinoma cell-line HCT-8/OR to oxaliplatin treatment through targeting ABCB1, ABCC2, ABCG2 and antiapoptosis pathways [48].
More investigations conducted in doxorubicin-resistant MCF-7 breast cancer cells partially supported our conclusion. Ectopic overexpression of miRNA-34a in MCF-7/doxorubicin cells could sensitize the resistant cells to chemotherapy by directly targeting Notch1, resulting in prevention of cell invasion using transwell assay, and induced early and late apoptosis and G1 phase arrest, as compared to negative control [37, 38]. Also, miRNA-34a mimics were recorded to increase the sensitivity and death of MDR-MCF-7 cells by targeting BCL-2, CCND1, and NOTCH1. The flow cytometric analysis of cell cycle distribution estimated a higher percentage of cells stayed in the G2/M phase, and the proportion of cells in the G0/G1 phase was lower than that of negative control-treated cells [32].
Contrary to our conclusion, Pu et al. reported that miRNA-34a-5p mimic-restoration sustained the multi-chemoresistance of MDR- SJSA-1 osteosarcoma cell lines via direct repression of angiotensin II type 1 receptor (AGTR1) [49], Delta-like ligand 1 (DLL1), the ligand of the Notch pathway, [50], and receptor tyrosine kinase CD117 [51].
Our results also showed an insignificant decrease in the percentage of cell population arrested in the S-phase (14.16 ± 0.23%) of the cell cycle as compared to negative control-treated MDA-MB-231/Dox cells (16.27 ± 0.96%). This observation was also confirmed by a study carried out by Niu et al. [52] using a model of human hepatocellular carcinoma SMMC7721 and MHCC97H cells.
Collectively, restoration of miRNA-34a-5p increases the sensitivity of the drug-resistant breast cancer cells via direct inhibition of the ABCC1 gene which is confirmed to has a role not only in mediating chemotherapeutic drugs exportation, but also in transporting a variety of substrates involved in inflammation and cellular signaling and proved in several different types of cancer to have a role in proliferation, clonogenic capacity, cell migration, and invasion, such as bioactive lipids sphingosine-1-phosphate, and lysophosphatidyl inositol [53]. Additionally, hence doxorubicin mediates its cell death action through a number of proposed models, including topoisomerase II poisoning, DNA adduct formation, oxidative stress, and ceramide overproduction, which are dependent on the corresponding doxorubicin dose [54], the maintained doxorubicin concentration as detected by influx assay will intercede the comparable cell death action.
Conclusion
In summary, the miRNA-34a-5p/ABCC1 axis has positive feedback effects on the sensitivity of MCF-7 and MDA-MB-231 cells to tamoxifen and doxorubicin, respectively, which is expected to be an important target for these chemotherapies in resistant breast cancer progression future studies. Totally, the current investigation introduced miRNA-34a-5p as positive regulators for ATP-binding cassette (ABC) transporter genes and oncogenic properties of drug-resistant breast cancer cells. MiRNA-34a could be used as a therapeutic molecule for reducing the developed drug resistance in breast cancer and the associated tumorigenic characteristics.
Data availability
All are available in the current research paper. For more required details, you could contact the corresponding author.
Abbreviations
- 3′UTR:
-
3′- Untranslated regions
- ABC:
-
ATP-binding cassette
- AGTR1:
-
Angiotensin II type 1 receptor
- ATCC:
-
American Type Culture Collection
- BC:
-
Breast cancer
- BCRP:
-
Breast cancer resistance protein
- DLL1:
-
Delta-like ligand 1
- DMEM:
-
Dulbecco’s Modified Eagle Medium
- DPBS:
-
Dulbecco’s phosphate-buffered saline
- EMT:
-
Epithelial to mesenchymal transition
- FBS:
-
Fetal Bovine Serum
- FLU:
-
Florescent and luminescent unit
- LDHA:
-
Lactate dehydrogenase A
- MCF-7/Tx:
-
MCF-7/ tamoxifen
- MDA-MB-231/Dox:
-
MDA-MB-231/Doxorubicin
- MDM4:
-
Murine Double Minute 4
- MDR:
-
Multidrug resistance
- MiRNA:
-
MicroRNA
- NC:
-
Negative control
- OAZ2:
-
Ornithine decarboxylase antizyme 2
- PBS:
-
Phosphate-buffered saline
- P-gp:
-
P-glycoprotein
- PI:
-
Propidium iodide
- RFU:
-
Relative Fluorescent Unit
- RLU:
-
Relative Light Unit
- SEM:
-
Standard error of mean
References
Xu Y, Gong M, Wang Y et al (2023) Global trends and forecasts of breast cancer incidence and deaths. Sci Data 10:334. https://doi.org/10.1038/s41597-023-02253-5
National Cancer Institute, surveillance, epidemiology and end result program (2023) (https://seer.cancer.gov/statfacts/html/breast.html).
Li J, Guo Y, Duan L, Hu X, Zhang X, Hu J, Huang L, He R, Hu Z, Luo W, Tan T, Huang R, Liao D, Zhu YS, Luo DX (2017) AKR1B10 promotes breast cancer cell migration and invasion via activation of ERK signaling. Oncotarget 8(20):33694–33703. https://doi.org/10.18632/oncotarget.16624
Kastl L, Brown I, Schofield AC (2012) miRNA-34a is associated with docetaxel resistance in human breast cancer cells. Breast Cancer Res Treat 131(2):445–454. https://doi.org/10.1007/s10549-011-1424-3
Pogribny IP, Filkowski JN, Tryndyak VP, Golubov A, Shpyleva SI, Kovalchuk O (2010) Alterations of microRNAs and their targets are associated with acquired resistance of MCF-7 breast cancer cells to cisplatin. Int J Cancer 127(8):1785–1794. https://doi.org/10.1002/ijc.25191
Garrido-Cano I, Pattanayak B, Adam-Artigues A, Lameirinhas A, Torres-Ruiz S, Tormo E, Cervera R, Eroles P (2022) MicroRNAs as a clue to overcome breast cancer treatment resistance. Cancer Metastasis Rev 41(1):77–105. https://doi.org/10.1007/s10555-021-09992-0
Hu W, Tan C, He Y, Zhang G, Xu Y, Tang J (2018) Functional miRNAs in breast cancer drug resistance. Onco Targets Ther 11:1529–1541. https://doi.org/10.2147/OTT.S152462
Morris K, Mattick J (2014) The rise of regulatory RNA. Nat Rev Genet 15:423–437. https://doi.org/10.1038/nrg3722
El-Toukhy SE, El-Daly SM, Kamel MM et al (2023) The diagnostic significance of circulating miRNAs and metabolite profiling in early prediction of breast cancer in Egyptian women. J Cancer Res Clin Oncol 149:5437–5451. https://doi.org/10.1007/s00432-022-04492-2
Zhang L, Liao Y, Tang L (2019) MicroRNA-34 family: a potential tumor suppressor and therapeutic candidate in cancer. J Exp Clin Cancer Res 38(1):53. https://doi.org/10.1186/s13046-019-1059-5
Yahya SM, Elsayed GH (2015) A summary for molecular regulations of miRNAs in breast cancer. Clin Biochem 48(6):388–396. https://doi.org/10.1016/j.clinbiochem.2014.12.013
Naghizadeh S, Mohammadi A, Duijf PHG, Baradaran B, Safarzadeh E, Cho WC, Mansoori B (2020) The role of miR-34 in cancer drug resistance. J Cell Physiol 235(10):6424–6440. https://doi.org/10.1002/jcp.29640
Imani S, Wu RC, Fu J (2018) MicroRNA-34 family in breast cancer: from research to therapeutic potential. J Cancer 9(20):3765–3775. https://doi.org/10.7150/jca.25576
Pan W, Chai B, Li L, Lu Z, Ma Z (2023) p53/MicroRNA-34 axis in cancer and beyond. Heliyon 9(4):e15155. https://doi.org/10.1016/j.heliyon.2023.e15155
Yahya SMM, Elmegeed GA, Mohamed MS, Mohareb RM, Abd-Elhalim MM, Elsayed GH (2018) The effect of newly synthesized heterosteroids on miRNA34a, 98, and 214 expression levels in MCF-7 breast cancer cells. Indian J Clin Biochem 33(3):328–333. https://doi.org/10.1007/s12291-017-0681-2
Liao R, Lin Y, Zhu L (2018) Molecular pathways involved in microRNA-mediated regulation of multidrug resistance. Mol Biol Rep 45(6):2913–2923. https://doi.org/10.1007/s11033-018-4358-6
Gomes BC, Honrado M, Armada A, Viveiros M, Rueff J, Rodrigues AS (2020) ABC efflux transporters and the circuitry of miRNAs: kinetics of expression in cancer drug resistance. Int J Mol Sci 21(8):2985. https://doi.org/10.3390/ijms21082985
He P, Liu X, Lou Y, Gong S, Cao L (2022) miR-34a-5p enhances the sensitivity of cervical cancer cells to oxaliplatin chemotherapy via targeting MDM4. Clin Exp Obstet Gynecol 49(2):54. https://doi.org/10.31083/j.ceog4902054
Pratama MY, Pascut D, Massi MN, Tiribelli C (2019) The role of microRNA in the resistance to treatment of hepatocellular carcinoma. Ann Transl Med 7(20):577. https://doi.org/10.21037/atm.2019.09.142
Kern F, Krammes L, Danz K, Diener C, Kehl T, Küchler O, Fehlmann T, Kahraman M, Rheinheimer S, Aparicio-Puerta E, Wagner S, Ludwig N, Backes C, Lenhof HP, von Briesen H, Hart M, Keller A, Meese E (2021) Validation of human microRNA target pathways enables evaluation of target prediction tools. Nucleic Acids Res 49(1):127–144. https://doi.org/10.1093/nar/gkaa1161
Yang X, Shang P, Yu B, Jin Q, Liao J, Wang L, Guo X (2021) Combination therapy with miR34a and doxorubicin synergistically inhibits Dox-resistant breast cancer progression via down-regulation of Snail through suppressing Notch/NF-κB and RAS/RAF/MEK/ERK signaling pathway. Acta Pharmaceutica Sinica B 11(9):2819–2834. https://doi.org/10.1016/j.apsb.2021.06.003
Rana NK, Singh P, Koch B (2019) CoCl2 simulated hypoxia induce cell proliferation and alter the expression pattern of hypoxia associated genes involved in angiogenesis and apoptosis. Biol Res 52(1):12. https://doi.org/10.1186/s40659-019-0221-z
Krisnamurti DG, Louisa M, Anggraeni E, Wanandi SI (2016) Drug efflux transporters are overexpressed in short-term tamoxifen-induced MCF7 breast cancer cells. Adv Pharmacol Sci 2016:6702424. https://doi.org/10.1155/2016/6702424
Yahya SMM, Fathy SA, El-Khayat ZA et al (2018) Possible role of microRNA-122 in modulating multidrug resistance of hepatocellular carcinoma. Ind J Clin Biochem 33:21–30. https://doi.org/10.1007/s12291-017-0651-8
Hamed AR, Yahya SMM, Nabih HK (2023) Anti-drug resistance, anti-inflammation, and anti-proliferation activities mediated by melatonin in doxorubicin-resistant hepatocellular carcinoma: in vitro investigations. Naunyn-Schmiedeberg’s Arch Pharmacol 396:1117–1128. https://doi.org/10.1007/s00210-023-02385-w
Nabih HK, Hamed AR, Yahya SMM (2023) Anti-proliferative effect of melatonin in human hepatoma HepG2 cells occurs mainly through cell cycle arrest and inflammation inhibition. Sci Rep 13:4396. https://doi.org/10.1038/s41598-023-31443-9
Dickson JR, Kruse C, Montagna DR, Finsen B, Wolfe MS (2013) Alternative polyadenylation and miR-34 family members regulate tau expression. J Neurochem 127(6):739–749. https://doi.org/10.1111/jnc.12437
Truksa J, Lee P, Beutler E (2007) The role of STAT, AP-1, E-box and TIEG motifs in the regulation of hepcidin by IL-6 and BMP-9: lessons from human HAMP and murine Hamp1 and Hamp2 gene promoters. Blood Cells Mol Dis 39(3):255–262. https://doi.org/10.1016/j.bcmd.2007.06.014
Punia R, Raina K, Agarwal R, Singh RP (2017) Acacetin enhances the therapeutic efficacy of doxorubicin in non-small-cell lung carcinoma cells. PLoS One 12(8):e0182870. https://doi.org/10.1371/journal.pone.0182870
Achari C, Winslow S, Ceder Y et al (2014) Expression of miR-34c induces G2/M cell cycle arrest in breast cancer cells. BMC Cancer 14:538. https://doi.org/10.1186/1471-2407-14-538
Park S, Kim H, Ji HW, Kim HW, Yun SH, Choi EH, Kim SJ (2019) Cold atmospheric plasma restores paclitaxel sensitivity to paclitaxel-resistant breast cancer cells by reversing expression of resistance-related genes. Cancers (Basel) 11(12):2011. https://doi.org/10.3390/cancers11122011
Li ZH, Weng X, Xiong QY, Tu JH, Xiao A, Qiu W, Gong Y, Hu EW, Huang S, Cao YL (2017) miR-34a expression in human breast cancer is associated with drug resistance. Oncotarget 8(63):106270–106282. https://doi.org/10.18632/oncotarget.22286
Han R, Zhao J, Lu L (2020) MicroRNA-34a expression affects breast cancer invasion in vitro and patient survival via downregulation of E2F1 and E2F3 expression. Oncol Rep 43(6):2062–2072. https://doi.org/10.3892/or.2020.7549
Kupcho K, Shultz J, Hurst R, Hartnett J, Zhou W, Machleidt T, Grailer J, Worzella T, Riss T, Lazar D, Cali JJ, Niles A (2019) A real-time, bioluminescent annexin V assay for the assessment of apoptosis. Apoptosis 24(1–2):184–197. https://doi.org/10.1007/s10495-018-1502-7
Li X, Zhao S, Fu Y, Zhang P, Zhang Z, Cheng J, Liu L, Jiang H (2021) miR-34a-5p functions as a tumor suppressor in head and neck squamous cell cancer progression by targeting Flotillin-2. Int J Biol Sci Int J Biol Sci 17(15):4327–4339. https://doi.org/10.7150/ijbs.64851
Suer İ, Kaya M, Özgür E (2021) The effect of miR-34a-5p and miR-145-5p ectopic expression on cell proliferation and target gene expression in the MDA-MB-231 cell line. Nam Kem Med J 9:166–173
Rui X, Zhao H, Xiao X, Wang L, Mo L, Yao Y (2018) MicroRNA-34a suppresses breast cancer cell proliferation and invasion by targeting Notch1. Exp Ther Med 16(6):4387–4392. https://doi.org/10.3892/etm.2018.6744
Li XJ, Ji MH, Zhong SL, Zha QB, Xu JJ, Zhao JH, Tang JH (2012) MicroRNA-34a modulates chemosensitivity of breast cancer cells to adriamycin by targeting Notch1. Arch Med Res 43(7):514–521. https://doi.org/10.1016/j.arcmed.2012.09.007
Mansoori B, Mohammadi A, Shirjang S, Baghbani E, Baradaran B (2016) Micro RNA 34a and Let-7a expression in human breast cancers is associated with apoptotic expression genes. Asian Pac J Cancer Prev 17(4):1887–1890. https://doi.org/10.7314/apjcp.2016.17.4.1887
Slabáková E, Culig Z, Remšík J, Souček K (2017) Alternative mechanisms of miR-34a regulation in cancer. Cell Death Dis 8(10):e3100. https://doi.org/10.1038/cddis.2017.495
Shao X, Zheng X, Ma D, Liu Y, Liu G (2021) Inhibition of lncRNA-NEAT1 sensitizes 5-Fu resistant cervical cancer cells through de-repressing the microRNA-34a/LDHA axis. Biosci Rep 41(7):BSR20200533. https://doi.org/10.1042/BSR20200533
Deng XJ, Zheng HL, Ke XQ, Deng M, Ma ZZ, Zhu Y, Cui YY (2021) Hsa-miR-34a-5p reverses multidrug resistance in gastric cancer cells by targeting the 3’-UTR of SIRT1 and inhibiting its expression. Cell Signal 84:110016. https://doi.org/10.1016/j.cellsig.2021.110016
Chou KY, Chang AC, Tsai TF, Lin YC, Chen HE, Ho CY, Chen PC, Hwang TI (2021) MicroRNA-34a-5p serves as a tumor suppressor by regulating the cell motility of bladder cancer cells through matrix metalloproteinase-2 silencing. Oncol Rep 45(3):911–920. https://doi.org/10.3892/or.2020.7910
Lv T, Song K, Zhang L, Li W, Chen Y, Diao Y, Yao Q, Liu P (2018) miRNA-34a decreases ovarian cancer cell proliferation and chemoresistance by targeting HDAC1. Biochem Cell Biol 96(5):663–671. https://doi.org/10.1139/bcb-2018-0031
Imani S, Wei C, Cheng J, Khan MA, Fu S, Yang L, Tania M, Zhang X, Xiao X, Zhang X, Fu J (2017) MicroRNA-34a targets epithelial to mesenchymal transition-inducing transcription factors (EMT-TFs) and inhibits breast cancer cell migration and invasion. Oncotarget 8(13):21362–21379. https://doi.org/10.18632/oncotarget.15214
Williams M, Cheng YY, Phimmachanh M, Winata P, van Zandwijk N, Reid G (2019) Tumour suppressor microRNAs contribute to drug resistance in malignant pleural mesothelioma by targeting anti-apoptotic pathways. Cancer Drug Resist 2(4):1193–1206. https://doi.org/10.20517/cdr.2019.41
Liu X, Luo X, Wu Y, Xia D, Chen W, Fang Z, Deng J, Hao Y, Yang X, Zhang T, Zhou L, Wu Y, Wang Q, Xu J, Hu X, Li L (2018) MicroRNA-34a attenuates paclitaxel resistance in prostate cancer cells via direct suppression of JAG1/Notch1 axis. Cell Physiol Biochem 50(1):261–276. https://doi.org/10.1159/000494004
Li Y, Gong P, Hou JX, Huang W, Ma XP, Wang YL, Li J, Cui XB, Li N (2018) miR-34a regulates multidrug resistance via positively modulating OAZ2 signaling in colon cancer cells. J Immunol Res 2018:7498514. https://doi.org/10.1155/2018/7498514
Pu Y, Zhao F, Li Y et al (2017) The miR-34a-5p promotes the multi-chemoresistance of osteosarcoma via repression of the AGTR1 gene. BMC Cancer 17:45. https://doi.org/10.1186/s12885-016-3002-x
Pu Y, Zhao F, Wang H, Cai S (2017) MiR-34a-5p promotes multi-chemoresistance of osteosarcoma through down-regulation of the DLL1 gene. Sci Rep 7:44218. https://doi.org/10.1038/srep44218
Pu Y, Zhao F, Wang H, Cai W, Gao J, Li Y, Cai S (2016) MiR-34a-5p promotes the multi-drug resistance of osteosarcoma by targeting the CD117 gene. Oncotarget 7(19):28420–28434. https://doi.org/10.18632/oncotarget.8546
Niu X, Wei N, Peng L, Li X, Zhang X, Wang C (2022) miR-34a-5p plays an inhibitory role in hepatocellular carcinoma by regulating target gene VEGFA. Malays J Pathol 44(1):39–52
Low FG, Shabir K, Brown JE, Bill RM, Rothnie AJ (2020) Roles of ABCC1 and ABCC4 in proliferation and migration of breast cancer cell lines. Int J Mol Sci 21(20):7664. https://doi.org/10.3390/ijms21207664
Yang F, Teves SS, Kemp CJ et al (2014) Doxorubicin, DNA torsion, and chromatin dynamics. Biochim. Biophys. Acta 1845:84–89. https://doi.org/10.1016/j.bbcan.2013.12.002
Acknowledgements
We'd like to express our gratitude to National Research Centre, Egypt, for the financial support of the current investigation under the grant number E120503. We would like to thank Dr. Mahmoud T. Abo-Elfadl, Cancer Biology and Genetics Laboratory, Centre of Excellence for Advanced Sciences, National Research Centre for providing technical guidance in this work.
Funding
Open access funding provided by The Science, Technology & Innovation Funding Authority (STDF) in cooperation with The Egyptian Knowledge Bank (EKB). This work was funded by the National Research Centre (Egypt)-project number (E120503).
Author information
Authors and Affiliations
Contributions
SMMY contributed to conceptualization, methodology, investigations, statistical analysis, validation, and funding acquisition; HKN contributed to investigations, statistical analysis, and interpretation of the results and wrote the manuscript; GHE, SIAM, and SMS contributed to visualization, investigations, validation, and editing; AME contributed to participated in the In Silico analysis. All authors reviewed and approved the submission. The authors declare that all data were generated in-house and that no paper mill was used.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Ethical approval
This investigation has complete compliance with ethical standards. This study does not involve human Participants or Animals.
Consent to participate
Not applicable.
Consent to publish
Not applicable.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Yahya, S.M.M., Nabih, H.K., Elsayed, G.H. et al. Restoring microRNA-34a overcomes acquired drug resistance and disease progression in human breast cancer cell lines via suppressing the ABCC1 gene. Breast Cancer Res Treat 204, 133–149 (2024). https://doi.org/10.1007/s10549-023-07170-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10549-023-07170-0