Abstract
Background
Breast cancer (BC) is a challenging disease and major cause of death amongst women worldwide who die due to tumor relapse or sidelong diseases. BC main complexity comes from the heterogeneous nature of breast tumors that demands customized treatments in the form of personalized medicine.
Review of the literature and discussion
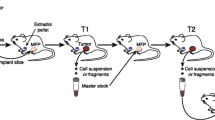
Spatiotemporally dynamic and heterogeneous nature of BC tumors is shaped by their clonal evolution and sub-clonal selections and shapes resistance to collective or group therapies that drives cancer recurrence and tumor metastasis. Personalized intervention promises to administer medications that selectively target each individual patient tumor and even further each colonized secondary tumor. Such personalized regimens will require creation of in vitro and in vivo models genuinely recapitulating characteristics of each tumor type as initiating platforms for two main purposes: to closely monitor the tumorigenic processes that shape tumor heterogeneity and evolution as the main driving forces behind tumor chemo-resistance and relapse, and subsequently to establish patient-specific preventive and therapeutic measures. While application of tumor modeling for personalized drug screening and design requires a separate review, here we discuss the personalized utilities of xenograft modeling in investigating BC tumor formation and progression toward metastasis. We will further elaborate on the impact of innovative technologies on personalized modeling of BC tumorigenicity at improved resolution.
Conclusion
Heterogeneous nature of each BC tumor requires personalized intervention implying that modeling breast tumors is inevitable for better disease understanding, detection and cure. Patient-derived xenografts are just the initiating piece of the puzzle for ideal management of breast cancer. Emerging technologies promise to model BC more personalized than before.



Similar content being viewed by others
Abbreviations
- AI:
-
Artificial intelligence
- BC:
-
Breast cancer
- BC-CSCs:
-
Breast cancer–cancer stem cells
- CSCs:
-
Cancer stem cells
- EMT:
-
Epithelial-to-mesenchymal transition
- GEMMs:
-
Genetically engineered mouse models
- iPS:
-
Induced pluripotent stem
- nGEMM:
-
Non-germ line GEMM
- NGS:
-
Next generation sequencing
- PDX:
-
Patient-derived xenograft
- PM:
-
Personalized medicine
- ts :
-
Tumor suppressor
References
Huxley J (1958) The biological aspects of cancer. Harcourt, Brace, New York
McGranahan N, Swanton C (2017) Clonal heterogeneity and tumor evolution: past, present, and the future. Cell 168(4):613–628. https://doi.org/10.1016/j.cell.2017.01.018
De Bruin EC, McGranahan N, Mitter R et al (2014) Spatial and temporal diversity in genomic instability processes defines lung cancer evolution. Science 346(6206):251–256. https://doi.org/10.1126/science.1253462
Mazor T, Pankov A, Song JS et al (2016) Intratumoral heterogeneity of the epigenome. Cancer Cell 29(4):440–451. https://doi.org/10.1016/j.ccell.2016.03.009
Martinez P, Birkbak NJ, Gerlinger M et al (2013) Parallel evolution of tumoursubclones mimics diversity between tumours. J Pathol 230(4):356–364. https://doi.org/10.1002/path.4214
Bombonati A, Sgroi DC (2011) The molecular pathology of breast cancer progression. J Pathol 223(2):308–318. https://doi.org/10.1002/path.2808
Polyak K (2007) Breast cancer: origins and evolution. J Clin Invest 117(11):3155–3163. https://doi.org/10.1172/JCI33295
Cancer Genome Atlas Network (2012) Comprehensive molecular portraits of human breast tumours. Nature 490(7418):61. https://doi.org/10.1038/nature11412
Curtis C, Shah SP, Chin SF et al (2012) The genomic and transcriptomic architecture of 2,000 breast tumoursreveals novel subgroups. Nature 486(7403):346. https://doi.org/10.1038/nature10983
Ali HR, Rueda OM, Chin SF et al (2014) Genome-driven integrated classification of breast cancer validated in over 7,500 samples. Genome Biol 15(8):431. https://doi.org/10.1186/s13059-014-0431-1
Malaney P, Nicosia SV, Davé V (2014) One mouse, one patient paradigm: New avatars of personalized cancer therapy. Cancer Lett 344(1):1–2. https://doi.org/10.1016/j.canlet.2013.10.010
Morton CL, Houghton PJ (2007) Establishment of human tumor xenografts in immunodeficient mice. Nat Protoc 2(2):247. https://doi.org/10.1038/nprot.2007.25
Jin K, Teng L, Shen Y et al (2010) Patient-derived human tumour tissue xenografts in immunodeficient mice: a systematic review. Clin Transl Oncol 12(7):473–480. https://doi.org/10.1007/s12094-010-0540-6
Cheon DJ, Orsulic S (2011) Mouse models of cancer. Annu Rev Pathol-Mech 6:95–119. https://doi.org/10.1146/annurev.pathol.3.121806.154244
Kersten K, de Visser KE, van Miltenburg MH et al (2016) Genetically engineered mouse models in oncology research and cancer medicine. EMBO Mol Med 9(2):137–153. https://doi.org/10.15252/emmm.201606857
Petrillo LA, Wolf DM, Kapoun AM et al (2012) Xenografts faithfully recapitulate breast cancer-specific gene expression patterns of parent primary breast tumors. Breast Cancer Res Treat 135(3):913–922. https://doi.org/10.1007/s10549-012-2226-y
Gardaneh M, Shojaei S, Kaviani A, Behnam B (2017) GDNF induces RET-SRC-HER2-dependent growth in trastuzumab-sensitive but SRC-independent growth in resistant breast tumor cells. Breast Cancer Res Treat 162(2):231–241. https://doi.org/10.1007/s10549-016-4078-3
Grinde MT, Skrbo N, Moestue SA et al (2014) Interplay of choline metabolites and genes in patient-derived breast cancer xenografts. Breast Cancer Res 16(1):R5. https://doi.org/10.1186/bcr3597
Bruna A, Rueda OM, Greenwood W et al (2016) A biobank of breast cancer explants with preserved intra-tumor heterogeneity to screen anticancer compounds. Cell 167(1):260–274. https://doi.org/10.1016/j.cell.2016.08.041
DeRose YS, Wang G, Lin YC et al (2011) Tumor grafts derived from women with breast cancer authentically reflect tumor pathology, growth, metastasis and disease outcomes. Nat Med 17(11):1514–1520. https://doi.org/10.1038/nm.2454
Zhang X, Claerhout S, Pratt A et al (2013) A renewable tissue resource of phenotypically stable, biologically and ethnically diverse, patient-derived human breast cancer xenograft models. Cancer Res 73:4885–4897. https://doi.org/10.1158/0008-5472.CAN-12-4081
Ding LI, Ellis MJ, Li S et al (2010) Genome remodeling in a basal-like breast cancer metastasis and xenograft. Nature 464(7291):999. https://doi.org/10.1038/nature08989
Nardella C, Lunardi A, Patnaik A et al (2011) The APL paradigm and the “co-clinical trial” project. Cancer Discov 1:108–116. https://doi.org/10.1158/2159-8290.CD-11-0061
Tentler JJ, Tan AC, Weekes CD et al (2012) Patient-derived tumour xenografts as models for oncology drug development. Nat Rev Clin Oncol 9(6):338–350. https://doi.org/10.1038/nrclinonc.2012.61
Eirew P, Steif A, Khattra J et al (2015) Dynamics of genomic clones in breast cancer patient xenografts at single-cell resolution. Nature 518(7539):422–426. https://doi.org/10.1038/nature13952
Reyal F, Guyader C, Decraene C et al (2012) Molecular profiling of patient-derived breast cancer xenografts. Breast Cancer Res 14:R11. https://doi.org/10.1186/bcr3095
Notta F, Mullighan CG, Wang JC et al (2011) Evolution of human BCR–ABL1 lymphoblastic leukaemia-initiating cells. Nature 469(7330):362–367. https://doi.org/10.1038/nature09733
Li S, Shen D, Shao J et al (2013) Endocrine-therapy-resistant ESR1 variants revealed by genomic characterization of breast-cancer-derived xenografts. Cell Reprod 4(6):1116–1130. https://doi.org/10.1016/j.celrep.2013.08.022
Ben-David U, Ha G, Tseng YY et al (2017) Patient-derived xenografts undergo mouse-specific tumor evolution. Nat Genet 49(11):1567–1575. https://doi.org/10.1038/ng.3967
Campbell PJ, Pleasance ED, Stephens PJ et al (2008) Subclonal phylogenetic structures in cancer revealed by ultra-deep sequencing. Proc Natl Acad Sci USA 105:13081–13086. https://doi.org/10.1073/pnas.0801523105
Lawson DA, Bhakta NR, Kessenbrock K et al (2015) Single-cell analysis reveals a stem-cell program in human metastatic breast cancer cells. Nature 526(7571):131–135. https://doi.org/10.1038/nature15260
Yang S, Zhang JJ, Huang XY (2012) Mouse models for tumor metastasis. Methods Mol Biol 928:221–228. https://doi.org/10.1007/978-1-62703-008-3_17
Gibson WJ, Hoivik EA, Halle MK et al (2016) The genomic landscape and evolution of endometrial carcinoma progression and abdominopelvic metastasis. Nat Genet 48(8):848–855. https://doi.org/10.1038/ng.3602
Diaz LA Jr., Bardelli A (2014) Liquid biopsies: genotyping circulating tumor DNA. J Clin Oncol 32(6):579. https://doi.org/10.1200/CO.2012.45.2011
Bettegowda C, Sausen M, Leary RJ et al (2014) Detection of circulating tumor DNA in early-and late-stage human malignancies. Sci Transl Med 6(224):224. https://doi.org/10.1126/scitranslmed.3007094
Thierry AR, Mouliere F, El Messaoudi S et al (2014) Clinical validation of the detection of KRAS and BRAF mutations from circulating tumor DNA. Nat Med 20(4):430. https://doi.org/10.1038/nm.3511
Jamal-Hanjani M, Wilson GA, Horswell S et al (2016) Detection of ubiquitous and heterogeneous mutations in cell-free DNA from patients with early-stage non-small-cell lung cancer. Ann Oncol 27(5):862–867. https://doi.org/10.1093/annonc/mdw037
Jamal-Hanjani M, Wilson GA, McGranahan N et al (2017) Tracking the evolution of non–small-cell lung cancer. N Engl J Med 376(22):2109–2121. https://doi.org/10.1056/NEJMoa1616288
Abbosh C, Birkbak NJ, Wilson GA et al (2017) Phylogenetic ctDNA analysis depicts early-stage lung cancer evolution. Nature 545(7655):446. https://doi.org/10.1038/nature22364
Diaz LA Jr, Williams RT, Wu J et al (2012) The molecular evolution of acquired resistance to targeted EGFR blockade in colorectal cancers. Nature 486(7404):537. https://doi.org/10.1038/nature11219
Misale S, Yaeger R, Hobor S et al (2012) Emergence of KRAS mutations and acquired resistance to anti-EGFR therapy in colorectal cancer. Nature 486(7404):532. https://doi.org/10.1038/nature11156
Spoerke JM, Gendreau S, Walter K et al (2016) Heterogeneity and clinical significance of ESR1 mutations in ER-positive metastatic breast cancer patients receiving fulvestrant. Nat Commun 7:11579. https://doi.org/10.1038/ncomms11579
Murtaza M, Dawson SJ, Tsui DW et al (2013) Non-invasive analysis of acquired resistance to cancer therapy by sequencing of plasma DNA. Nature 497(7447):108. https://doi.org/10.1038/nature12065
Leary RJ, Sausen M, Kinde I et al (2012) Detection of chromosomal alterations in the circulation of cancer patients with whole-genome sequencing. Sci Transl Med 4(162):154–162. https://doi.org/10.1126/scitranslmed.3004742
Shoda K, Ichikawa D, Fujita Y et al (2017) Monitoring the HER2 copy number status in circulating tumor DNA by droplet digital PCR in patients with gastric cancer. Gastric Cancer 20(1):126–135. https://doi.org/10.1007/s10120-017-0715-8
Garcia-Murillas I, Schiavon G, Weigelt B, Ng C, Hrebien S, Cutts RJ, Cheang M, Osin P, Nerurkar A, Kozarewa I, Garrido JA, Dowsett M, Reis-Filho JS, Smith IE, Turner NC (2105) Mutation tracking in circulating tumor DNA predicts relapse in early breast cancer. Sci Transl Med 26(302):302ra133. https://doi.org/10.1126/scitranslmed.aab0021
Yang P, Abo R, Liu Ch, Chen Z, Wu H, Cui J, Yandava Ch, Baily ST, Balch C, Gulcher JR, Chittenden TW (2017) Novel feature selection strategies for enhanced predictive modeling and deep learning in the biosciences [abstract]. In: Proceedings of the American Association for Cancer Research Annual Meeting 2017; 2017 Apr 1–5; Washington DC Philadelphia (PA): AACR; Cancer Res 77(13 Suppl):Abstract nr 4539. https://doi.org/10.1158/1538-7445.AM2017-4539
Caravagna G, Giarratano Y, Ramazzotti D et al (2018) Detecting repeated cancer evolution from multi-region tumor sequencing data. Nat Methods 15(9):707. https://doi.org/10.1038/s41592-018-0108-x
Way GP, Sanchez-Vega F, La K et al (2018) Machine learning detects pan-cancer Ras pathway activation in the cancer genome atlas. Cell Rep 23:172–180. https://doi.org/10.1016/j.celrep.2018.03.046
Li J, Chen L, Zhang YH et al (2018) A computational method for classifying different human tissues with quantitatively tissue-specific expressed genes. Genes 9(9):449. https://doi.org/10.3390/genes9090449
Zinn PO, Singh SK, Kotrotsou A et al (2018) Validation study: conserved magnetic resonance radiomic appearance of periostin-expressing glioblastoma in patients and xenograft models. Clin Cancer Res 24(24):6288–6299. https://doi.org/10.1158/1078-0432.CCR-17-3420
Wang D, Li JR, Zhang YH et al (2018) Identification of differentially expressed genes between original breast cancer and xenograft using machine learning algorithms. Genes 9(3):155. https://doi.org/10.3390/genes9030155
Jeanquartier F, Jean-Quartier C, Cemernek D et al (2016) In silico modeling for tumor growth visualization. BMC Syst Biol 10(1):59. https://doi.org/10.1186/s12918-016-0318-8
Drost J, Clevers H (2018) Organoids in cancer research. Nat Rev Cancer 24:1. https://doi.org/10.1038/s41568-018-0007-6
Vlachogiannis G, Hedayat S, Vatsiou A et al (2018) Patient-derived organoids model treatment response of metastatic gastrointestinal cancers. Science 359 (6378):920–926. https://doi.org/10.1126/science.aao2774
Byrne AT, Alférez DG, Amant F, Annibali D, Arribas J, Biankin AV, Bruna A, Budinská E, Caldas C, Chang DK (2017) Interrogating open issues in cancer precision medicine with patient-derived xenografts. Nat Rev Cancer 17(4):254. https://doi.org/10.1038/nrc.2016.140
Wang H, Russa ML, Qi LS (2016) CRISPR/Cas9 in genome editing and beyond. Annu Rev Biochem 85:227–264. https://doi.org/10.1146/annurev-biochem-060815-014607
Choi PS, Meyerson M (2014) Targeted genomic rearrangements using CRISPR/Cas technology. Nat Commun 5:3728. https://doi.org/10.1038/ncomms4728
Chen C, Liu Y, Rappaport AR et al (2014) MLL3 is a haploinsufficient 7q tumor suppressor in acute myeloid leukemia. Cancer Cell 25(5):652–665. https://doi.org/10.1016/j.ccr.2014.03.016
Torres R, Martin MC, Garcia A et al (2014) Engineering human tumour-associated chromosomal translocations with the RNA-guided CRISPR–Cas9 system. Nat Commun 5:3964. https://doi.org/10.1038/ncomms4964
Guernet A, Mungamuri SK, Cartier D et al (2016) CRISPR-barcoding for intratumor genetic heterogeneity modeling and functional analysis of oncogenic driver mutations. Mol Cell 63(3):526–538. https://doi.org/10.1016/j.molcel.2016.06.017
Frieda KL, Linton JM, Hormoz S et al (2017) Synthetic recording and in situ readout of lineage information in single cells. Nature 541(7635):107–111. https://doi.org/10.1038/nature20777
Engelman JA, Chen L, Tan X et al (2008) Effective use of PI3K and MEK inhibitors to treat mutant Kras G12D and PIK3CA H1047R murine lung cancers. Nat Med 14(12):1351. https://doi.org/10.1038/nm.1890
Chen Z, Cheng K, Walton Z et al (2012) A murine lung cancer co-clinical trial identifies genetic modifiers of therapeutic response. Nature 483(7391):613. https://doi.org/10.1038/nature10937
Yang H, Wang H, Shivalila CS et al (2013) One-step generation of mice carrying reporter and conditional alleles by CRISPR/Cas-mediated genome engineering. Cell 154(6):1370–1379. https://doi.org/10.1016/j.cell.2013.08.022
Malina A, Mills JR, Cencic R et al (2013) Repurposing CRISPR/Cas9 for in situ functional assays. Genes Dev 27(23):2602–2614. https://doi.org/10.1101/gad.227132.113
Heckl D, Kowalczyk MS, Yudovich D et al (2014) Generation of mouse models of myeloid malignancy with combinatorial genetic lesions using CRISPR-Cas9 genome editing. Nat Biotechnol 32(9):941. https://doi.org/10.1038/nbt.2951
Xue W, Chen S, Yin H et al (2014) CRISPR-mediated direct mutation of cancer genes in the mouse liver. Nature 514(7522):380. https://doi.org/10.1038/nature13589
Jackson EL, Willis N, Mercer K et al (2001) Analysis of lung tumor initiation and progression using conditional expression of oncogenic K-ras. Genes Dev 15(24):3243–3248. https://doi.org/10.1101/gad.943001
Sánchez-Rivera FJ, Papagiannakopoulos T, Romero R et al (2014) Rapid modelling of cooperating genetic events in cancer through somatic genome editing. Nature 516(7531):428. https://doi.org/10.1038/nature13906
Blasco RB, Karaca E, Ambrogio C et al (2014) Simple and rapid in vivo generation of chromosomal rearrangements using CRISPR/Cas9 technology. Cell Rep 9(4):1219–1227. https://doi.org/10.1016/j.celrep.2014.10.051
Soda M, Choi YL, Enomoto M et al (2007) Identification of the transforming EML4–ALK fusion gene in non-small-cell lung cancer. Nature 448(7153):561. https://doi.org/10.1038/nature05945
Maddalo D, Manchado E, Concepcion CP et al (2014) In vivo engineering of oncogenic chromosomal rearrangements with the CRISPR/Cas9 system. Nature 516(7531):423. https://doi.org/10.1038/nature13902
Hockemeyer D, Jaenisch R (2016) Induced pluripotent stem cells meet genome editing. Cell Stem Cell 18(5):573–586. https://doi.org/10.1016/j.stem.2016.04.013
Acknowledgements
This study was financially supported by a Grant (502) from NIGEB. We thank all staff in Stem Cells Lab in NIGEB for their cooperation. Mossa Gardaneh would like to dedicate his share of this study to his beloved hometown Benis for all the inspirations he has received from in his lifetime.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
This article does not contain any studies with human participants or animals. The study was approved by the institutional review board for a retrospective chart review.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Afzali, F., Akbari, P., Naderi-Manesh, H. et al. The next generation personalized models to screen hidden layers of breast cancer tumorigenicity. Breast Cancer Res Treat 175, 277–286 (2019). https://doi.org/10.1007/s10549-019-05159-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10549-019-05159-2