Abstract
Purpose
We aimed to establish the spectrum of BRCA1/2 mutations among the breast cancer (BC) patients from the Republic of Macedonia.
Methods
We used targeted next-generation sequencing (NGS), Sanger DNA sequencing, and multiplex ligation probe amplification analysis (MLPA) to search for point mutations and deletions/duplications involving BRCA1 and BRCA2-coding regions.
Results
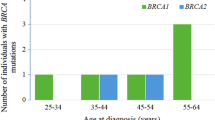
We have analyzed a total of 313 BC patients, enriched for family history of cancer, early age of onset and bilateral and/or triple negative (TN) BC. A total of 26 pathogenic mutations were observed in 49 unrelated BC patients (49/313, 15.7%). BRCA2 mutations (27/49, 55.1%) were more common than BRCA1 mutations (22/49, 44.9%). We identified five novel point mutations, one in BRCA1 (c.4352_4356delA) and four in BRCA2 (c.151G>T, c.4707_4708delCA, c.7811_7814delTGTG, and c.9304_9305delG), as well as two novel deletions involving parts of the BRCA1 gene (c.81−?_593+?del and c.5470−?_5530+?del). The most common mutations were c.181T>G, c.5266dupC, and c.3700_3704del5 in BRCA1 and c.7879A>T, c.8317_8330del14 and c.5722_5723delCT in BRCA2 gene. Thus far, BRCA2 c.7879A>T and c.8317_8330del14 mutations have been described in several isolated cases; however, our study is the first one showing that they have a founder effect among Macedonian population. Nine recurrent mutations account for 65.3% of all of the detected mutations allowing for implementation of a fast first-step BRCA1/2 mutational screening strategy in our country.
Conclusion
This study provides a comprehensive view of known and novel BRCA1/2 mutations in BC patients from the Republic of Macedonia and contributes to the global spectrum of BRCA1/2 mutations in breast cancer.

Similar content being viewed by others
References
Miki Y, Swensen J, Shattuck-Eidens D, Futreal PA, Harshman K, Tavtigian S, Liu Q, Cochran C, Bennett LM, Ding W et al (1994) A strong candidate for the breast and ovarian cancer susceptibility gene BRCA1. Science 266(5182):66–71
Wooster R, Bignell G, Lancaster J, Swift S, Seal S, Mangion J, Collins N, Gregory S, Gumbs C, Micklem G (1995) Identification of the breast cancer susceptibility gene BRCA2. Nature 378(6559):789–792. https://doi.org/10.1038/378789a0
Antoniou A, Pharoah PD, Narod S, Risch HA, Eyfjord JE, Hopper JL, Loman N, Olsson H, Johannsson O, Borg A, Pasini B, Radice P, Manoukian S, Eccles DM, Tang N, Olah E, Anton-Culver H, Warner E, Lubinski J, Gronwald J, Gorski B, Tulinius H, Thorlacius S, Eerola H, Nevanlinna H, Syrjakoski K, Kallioniemi OP, Thompson D, Evans C, Peto J, Lalloo F, Evans DG, Easton DF (2003) Average risks of breast and ovarian cancer associated with BRCA1 or BRCA2 mutations detected in case Series unselected for family history: a combined analysis of 22 studies. Am J Hum Genet 72(5):1117–1130. https://doi.org/10.1086/375033
Ford D, Easton DF, Bishop DT, Narod SA, Goldgar DE (1994) Risks of cancer in BRCA1-mutation carriers. Breast Cancer Linkage Consortium. Lancet 343(8899):692–695
Easton DF, Ford D, Bishop DT (1995) Breast and ovarian cancer incidence in BRCA1-mutation carriers. Breast Cancer Linkage Consortium. Am J Hum Genet 56(1):265–271
Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, Kondrashov AS, Sunyaev SR (2010) A method and server for predicting damaging missense mutations. Nat Methods 7(4):248–249. https://doi.org/10.1038/nmeth0410-248
Kumar P, Henikoff S, Ng PC (2009) Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat Protoc 4(7):1073–1081. https://doi.org/10.1038/nprot.2009.86
Choi Y, Sims GE, Murphy S, Miller JR, Chan AP (2012) Predicting the functional effect of amino acid substitutions and indels. PLoS ONE 7(10):e46688. https://doi.org/10.1371/journal.pone.0046688
Reva B, Antipin Y, Sander C (2011) Predicting the functional impact of protein mutations: application to cancer genomics. Nucleic Acids Res 39(17):e118. https://doi.org/10.1093/nar/gkr407
Schwarz JM, Cooper DN, Schuelke M, Seelow D (2014) MutationTaster2: mutation prediction for the deep-sequencing age. Nat Methods 11(4):361–362
Yates CM, Filippis I, Kelley LA, Sternberg MJ (2014) SuSPect: enhanced prediction of single amino acid variant (SAV) phenotype using network features. J Mol Biol 426(14):2692–2701
Mi H, Poudel S, Muruganujan A, Casagrande JT, Thomas PD (2016) PANTHER version 10: expanded protein families and functions, and analysis tools. Nucleic Acids Res 44(D1):17
Desmet FO, Hamroun D, Lalande M, Collod-Beroud G, Claustres M, Beroud C (2009) Human Splicing Finder: an online bioinformatics tool to predict splicing signals. Nucleic Acids Res 37(9):e67. https://doi.org/10.1093/nar/gkp215
Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, Grody WW, Hegde M, Lyon E, Spector E, Voelkerding K, Rehm HL (2015) Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med 17(5):405–424. https://doi.org/10.1038/gim.2015.30
Maleva I, Madjunkova S, Bozhinovski G, Smickova E, Kondov G, Spiroski Z, Arsovski A, Plaseska-Karanfilska D (2012) Genetic variation of the brca1 and brca2 genes in macedonian patients. Balkan J Med Genet 15(Suppl):81–85. https://doi.org/10.2478/v10034-012-0025-8
Dobricic J, Krivokuca A, Brotto K, Malisic E, Radulovic S, Brankovic-Magic M (2013) Serbian high-risk families: extensive results on BRCA mutation spectra and frequency. J Hum Genet 58(8):501–507
Dodova RI, Mitkova AV, Dacheva DR, Hadjo LB, Vlahova AI, Hadjieva MST, Valev SS, Caulevska MM, Popova SD, Popov IE, Dikov TI, Sedloev TA, Ionkov AS, Timcheva KV, Christova SL, Kremensky IM, Mitev VI, Kaneva RP (2015) Spectrum and frequencies of BRCA1/2 mutations in Bulgarian high risk breast cancer patients. BMC Cancer 15(523):015–1516
Konstantopoulou I, Rampias T, Ladopoulou A, Koutsodontis G, Armaou S, Anagnostopoulos T, Nikolopoulos G, Kamakari S, Nounesis G, Stylianakis A, Karanikiotis C, Razis E, Gogas H, Keramopoulos A, Gaki V, Markopoulos C, Skarlos D, Pandis N, Bei T, Arzimanoglou I, Fountzilas G, Yannoukakos D (2008) Greek BRCA1 and BRCA2 mutation spectrum: two BRCA1 mutations account for half the carriers found among high-risk breast/ovarian cancer patients. Breast Cancer Res Treat 107(3):431–441
Konstantopoulou I, Tsitlaidou M, Fostira F, Pertesi M, Stavropoulou AV, Triantafyllidou O, Tsotra E, Tsiftsoglou AP, Tsionou C, Droufakou S, Dimitrakakis C, Fountzilas G, Yannoukakos D (2014) High prevalence of BRCA1 founder mutations in Greek breast/ovarian families. Clin Genet 85(1):36–42
Levanat S, Musani V, Cvok ML, Susac I, Sabol M, Ozretic P, Car D, Eljuga D, Eljuga L (2012) Three novel BRCA1/BRCA2 mutations in breast/ovarian cancer families in Croatia. Gene 498(2):169–176
Krajc M, Teugels E, Zgajnar J, Goelen G, Besic N, Novakovic S, Hocevar M, De Greve J (2008) Five recurrent BRCA1/2 mutations are responsible for cancer predisposition in the majority of Slovenian breast cancer families. BMC Med Genet 9(83):1471–2350
Stegel V, Krajc M, Zgajnar J, Teugels E, De Greve J, Hocevar M, Novakovic S (2011) The occurrence of germline BRCA1 and BRCA2 sequence alterations in Slovenian population. BMC Med Genet 12:9. https://doi.org/10.1186/1471-2350-12-9
Gorski B, Byrski T, Huzarski T, Jakubowska A, Menkiszak J, Gronwald J, Pluzanska A, Bebenek M, Fischer-Maliszewska L, Grzybowska E, Narod SA, Lubinski J (2000) Founder mutations in the BRCA1 gene in polish families with breast-ovarian cancer. Am J Hum Genet 66(6):1963–1968
Gorski B, Jakubowska A, Huzarski T, Byrski T, Gronwald J, Grzybowska E, Mackiewicz A, Stawicka M, Bebenek M, Sorokin D, Fiszer-Maliszewska L, Haus O, Janiszewska H, Niepsuj S, Gozdz S, Zaremba L, Posmyk M, Pluzanska M, Kilar E, Czudowska D, Wasko B, Miturski R, Kowalczyk JR, Urbanski K, Szwiec M, Koc J, Debniak B, Rozmiarek A, Debniak T, Cybulski C, Kowalska E, Toloczko-Grabarek A, Zajaczek S, Menkiszak J, Medrek K, Masojc B, Mierzejewski M, Narod SA, Lubinski J (2004) A high proportion of founder BRCA1 mutations in Polish breast cancer families. Int J Cancer 110(5):683–686
Kluska A, Balabas A, Paziewska A, Kulecka M, Nowakowska D, Mikula M, Ostrowski J (2015) New recurrent BRCA1/2 mutations in Polish patients with familial breast/ovarian cancer detected by next generation sequencing. BMC Med Genom 8(19):015–0092
Machackova E, Foretova L, Lukesova M, Vasickova P, Navratilova M, Coene I, Pavlu H, Kosinova V, Kuklova J, Claes K (2008) Spectrum and characterisation of BRCA1 and BRCA2 deleterious mutations in high-risk Czech patients with breast and/or ovarian cancer. BMC Cancer 8(140):1471–2407
Spitzer E, Abbaszadegan MR, Schmidt F, Hauser A, Buwitt U, Lauter FR, Potschick K, Krocker J, Elling D, Grosse R (2000) Detection of BRCA1 and BRCA2 mutations in breast cancer families by a comprehensive two-stage screening procedure. Int J Cancer 85(4):474–481
Cecener G, Egeli U, Tunca B, Erturk E, Ak S, Gokgoz S, Tasdelen I, Tezcan G, Demirdogen E, Bayram N, Avci N, Evrensel T (2014) BRCA1/2 germline mutations and their clinical importance in Turkish breast cancer patients. Cancer Investig 32(8):375–387
Stegel V, Krajc M, Zgajnar J, Teugels E, De Greve J, Hocevar M, Novakovic S (2011) The occurrence of germline BRCA1 and BRCA2 sequence alterations in Slovenian population. BMC Med Genet 12(9):1471–2350
van der Hout AH, van den Ouweland AM, van der Luijt RB, Gille HJ, Bodmer D, Bruggenwirth H, Mulder IM, van der Vlies P, Elfferich P, Huisman MT, ten Berge AM, Kromosoeto J, Jansen RP, van Zon PH, Vriesman T, Arts N, Lange MB, Oosterwijk JC, Meijers-Heijboer H, Ausems MG, Hoogerbrugge N, Verhoef S, Halley DJ, Vos YJ, Hogervorst F, Ligtenberg M, Hofstra RM (2006) A DGGE system for comprehensive mutation screening of BRCA1 and BRCA2: application in a Dutch cancer clinic setting. Hum Mutat 27(7):654–666
Yang H, Jeffrey PD, Miller J, Kinnucan E, Sun Y, Thoma NH, Zheng N, Chen PL, Lee WH, Pavletich NP (2002) BRCA2 function in DNA binding and recombination from a BRCA2-DSS1-ssDNA structure. Science 297(5588):1837–1848
Caux-Moncoutier V, Castera L, Tirapo C, Michaux D, Remon MA, Lauge A, Rouleau E, De Pauw A, Buecher B, Gauthier-Villars M, Viovy JL, Stoppa-Lyonnet D, Houdayer C (2011) EMMA, a cost- and time-effective diagnostic method for simultaneous detection of point mutations and large-scale genomic rearrangements: application to BRCA1 and BRCA2 in 1,525 patients. Hum Mutat 32(3):325–334
Houdayer C, Caux-Moncoutier V, Krieger S, Barrois M, Bonnet F, Bourdon V, Bronner M, Buisson M, Coulet F, Gaildrat P, Lefol C, Leone M, Mazoyer S, Muller D, Remenieras A, Revillion F, Rouleau E, Sokolowska J, Vert JP, Lidereau R, Soubrier F, Sobol H, Sevenet N, Bressac-de Paillerets B, Hardouin A, Tosi M, Sinilnikova OM, Stoppa-Lyonnet D (2012) Guidelines for splicing analysis in molecular diagnosis derived from a set of 327 combined in silico/in vitro studies on BRCA1 and BRCA2 variants. Hum Mutat 33(8):1228–1238
Guidugli L, Pankratz VS, Singh N, Thompson J, Erding CA, Engel C, Schmutzler R, Domchek S, Nathanson K, Radice P, Singer C, Tonin PN, Lindor NM, Goldgar DE, Couch FJ (2013) A classification model for BRCA2 DNA binding domain missense variants based on homology-directed repair activity. Cancer Res 73(1):265–275
Michailidou K, Hall P, Gonzalez-Neira A, Ghoussaini M, Dennis J, Milne RL, Schmidt MK, Chang-Claude J, Bojesen SE, Bolla MK, Wang Q, Dicks E, Lee A, Turnbull C, Rahman N, Fletcher O, Peto J, Gibson L, Dos Santos Silva I, Nevanlinna H, Muranen TA, Aittomaki K, Blomqvist C, Czene K, Irwanto A, Liu J, Waisfisz Q, Meijers-Heijboer H, Adank M, van der Luijt RB, Hein R, Dahmen N, Beckman L, Meindl A, Schmutzler RK, Muller-Myhsok B, Lichtner P, Hopper JL, Southey MC, Makalic E, Schmidt DF, Uitterlinden AG, Hofman A, Hunter DJ, Chanock SJ, Vincent D, Bacot F, Tessier DC, Canisius S, Wessels LF, Haiman CA, Shah M, Luben R, Brown J, Luccarini C, Schoof N, Humphreys K, Li J, Nordestgaard BG, Nielsen SF, Flyger H, Couch FJ, Wang X, Vachon C, Stevens KN, Lambrechts D, Moisse M, Paridaens R, Christiaens MR, Rudolph A, Nickels S, Flesch-Janys D, Johnson N, Aitken Z, Aaltonen K, Heikkinen T, Broeks A, Veer LJ, van der Schoot CE, Guenel P, Truong T, Laurent-Puig P, Menegaux F, Marme F, Schneeweiss A, Sohn C, Burwinkel B, Zamora MP, Perez JI, Pita G, Alonso MR, Cox A, Brock IW, Cross SS, Reed MW, Sawyer EJ, Tomlinson I, Kerin MJ, Miller N, Henderson BE, Schumacher F, Le Marchand L, Andrulis IL, Knight JA, Glendon G, Mulligan AM, Lindblom A, Margolin S, Hooning MJ, Hollestelle A, van den Ouweland AM, Jager A, Bui QM, Stone J, Dite GS, Apicella C, Tsimiklis H, Giles GG, Severi G, Baglietto L, Fasching PA, Haeberle L, Ekici AB, Beckmann MW, Brenner H, Muller H, Arndt V, Stegmaier C, Swerdlow A, Ashworth A, Orr N, Jones M, Figueroa J, Lissowska J, Brinton L, Goldberg MS, Labreche F, Dumont M, Winqvist R, Pylkas K, Jukkola-Vuorinen A, Grip M, Brauch H, Hamann U, Bruning T, Radice P, Peterlongo P, Manoukian S, Bonanni B, Devilee P, Tollenaar RA, Seynaeve C, van Asperen CJ, Jakubowska A, Lubinski J, Jaworska K, Durda K, Mannermaa A, Kataja V, Kosma VM, Hartikainen JM, Bogdanova NV, Antonenkova NN, Dork T, Kristensen VN, Anton-Culver H, Slager S, Toland AE, Edge S, Fostira F, Kang D, Yoo KY, Noh DY, Matsuo K, Ito H, Iwata H, Sueta A, Wu AH, Tseng CC, Van Den Berg D, Stram DO, Shu XO, Lu W, Gao YT, Cai H, Teo SH, Yip CH, Phuah SY, Cornes BK, Hartman M, Miao H, Lim WY, Sng JH, Muir K, Lophatananon A, Stewart-Brown S, Siriwanarangsan P, Shen CY, Hsiung CN, Wu PE, Ding SL, Sangrajrang S, Gaborieau V, Brennan P, McKay J, Blot WJ, Signorello LB, Cai Q, Zheng W, Deming-Halverson S, Shrubsole M, Long J, Simard J, Garcia-Closas M, Pharoah PD, Chenevix-Trench G, Dunning AM, Benitez J, Easton DF (2013) Large-scale genotyping identifies 41 new loci associated with breast cancer risk. Nat Genet 45(4):353–361
Acknowledgements
We cordially thank all patients and clinicians who participated in sampling and documentation.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional research committees and with the 1964 Helsinki Declaration and its later amendments or comparable ethical standards.
Informed consent
Informed consent was obtained from all individual participants included in the study.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Jakimovska, M., Maleva Kostovska, I., Popovska-Jankovic, K. et al. BRCA1 and BRCA2 germline variants in breast cancer patients from the Republic of Macedonia. Breast Cancer Res Treat 168, 745–753 (2018). https://doi.org/10.1007/s10549-017-4642-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10549-017-4642-5