Abstract
Computational models lie at the intersection of basic neuroscience and healthcare applications because they allow researchers to test hypotheses in silico and predict the outcome of experiments and interactions that are very hard to test in reality. Yet, what is meant by “computational model” is understood in many different ways by researchers in different fields of neuroscience and psychology, hindering communication and collaboration. In this review, we point out the state of the art of computational modeling in Electroencephalography (EEG) and outline how these models can be used to integrate findings from electrophysiology, network-level models, and behavior. On the one hand, computational models serve to investigate the mechanisms that generate brain activity, for example measured with EEG, such as the transient emergence of oscillations at different frequency bands and/or with different spatial topographies. On the other hand, computational models serve to design experiments and test hypotheses in silico. The final purpose of computational models of EEG is to obtain a comprehensive understanding of the mechanisms that underlie the EEG signal. This is crucial for an accurate interpretation of EEG measurements that may ultimately serve in the development of novel clinical applications.
Similar content being viewed by others
Introduction
Electroencephalography (EEG) has applications in many fields, spanning from basic neuroscientific research to clinical domains. However, despite the technological advances in recording precision, the full potential of EEG is currently not being exploited. One possible way to do so is to use computational models in order to integrate findings from electrophysiology, network-level models (the level of neuroimaging), and behavior (Franceschiello et al. 2018, 2019). The following review has been conceived with the specific goal of targeting a non-expert audience. Indeed this does not constitute an exhaustive review, but we believe that structuring the contents along spatial scales might facilitate the understanding of this broad topic. Furthermore, this review does not cover Brain Computer Interfaces (BCIs), first as they constitute an independent topic itself at the interface between engineering and neuroscience (Bulárka and Gontean 2016). Secondly, the vast majority of BCI works relies on a statistical model of the neural signal, often combined with machine learning approaches, and thus lie outside the scope of this review. Here, our aim is to discuss computational models that integrate cellular behavior at different spatial scales and make explicit links to EEG empirical data. We refer readers interested in BCI to Wolpaw et al. (2000), Schiff (2012), Fouad et al. (2015), Bulárka and Gontean (2016), and for BCI-based EEG paradigms to Abiri et al. (2019).
A model is defined in terms of a set of equations which describe the relationships between variables. Importantly, models exist for different spatial scales (Varela et al. 2001; Deco et al. 2008; Nunez et al. 2019), spanning from the single cell spike train up to macroscopic oscillations. The equations are used to simulate how each variable changes over time, or, in rare cases, to find analytical solutions for the relationships among the variables. The dynamics of the resulting time series are also influenced by a set of parameters, which can either be estimated from available data—for example, a model which simulates the firing of a certain neuron type could contain a time constant estimated from recordings on that type of neuron in rodents—or its value can be varied systematically in an exploratory manner. The goal is to produce time series of variables that can be compared to real data. In particular, one can simulate perturbations to brain activity, be it sensory stimulation, a therapeutic intervention like deep brain stimulation (DBS) or a drug, or a structural change due to the onset of a pathology, like neurodegeneration or a lesion, and predict what would be the resulting alterations observed in neural and clinical data.
An important application of EEG models is in the clinical domain. Psychiatric and neurological disorders impact a growing portion of the population, both as patients and caregivers, and with an enormous cost—both economical and humanitarian—to healthcare systems worldwide (Steel et al. 2014; Vigo et al. 2016; Feigin et al. 2019). One of the main obstacles in advancing patient care is the lack of individual diagnosis, prognosis, and treatment planning (Wium-Andersen et al. 2017). Computational models can be adapted to the individual by setting their parameters according to available data (i.e. either setting the parameter directly, if it is measurable, or looking for the parameter value which results in time series whose dynamics match recorded data). The adjusted parameter(s) can then be related to clinical markers, symptoms, and behavior, allowing for example to discriminate between pathologies. Using models in this personalized manner could provide additional diagnostic features in the form of model parameters and model output, eventually assisting clinicians in diagnosis and treatment planning.
Another obstacle is a general lack of scientific knowledge of disease mechanisms, including the mechanisms by which therapies exert their effect. As an example, DBS is a highly effective treatment for advanced Parkinsonism, in which electric pulses are delivered directly to certain deep brain structures via permanently implanted electrodes. Yet, it is largely unknown how exactly the applied stimulation manages to suppress motor symptoms such as tremor (Chiken and Nambu 2016). This is also because the way in which motor symptoms result from the degradation of dopaminergic neurons in the substantia nigra is not fully understood (McGregor and Nelson 2019). Besides animal models—which have their own ethical issues—in silico models are an indispensable tool for understanding brain disorders. Combining data available from a patient or group of patients with knowledge and hypotheses about mechanisms, a model can be generated which can help test these hypotheses.
Last but not least, models are much cheaper than animal testing or clinical trials. While models will not replace these approaches—at least not in the foreseeable future—they could help to formulate more specific hypotheses and thus, lead to smaller-scale experiments.
Collecting invasive data is not generally possible in humans. EEG (Nunez et al. 2006; Schomer and Lopes Da Silva 2012; Biasiucci et al. 2019) is an extremely versatile technology which allows non-invasive recording of neural activity in behaving humans. EEG is a cheap and portable technology, particularly compared to (f)MRI and MEG. Apart from these cost-efficiency considerations, EEG, like MEG, is a direct measure of the electromagnetic fields generated by the brain, and allows millisecond-precision recordings, thus giving access to rich aspects of brain function which can inform models in a way that e.g. fMRI cannot (see “Electroencephalography” section for more details). In general, using different complementary sources of data to construct and validate a model will lead to better model predictions, as each recording technique has its own strengths and weaknesses, and a multimodal approach can balance them.
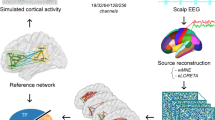
In our opinion, there are mostly two reasons why EEG has not been used more extensively in modeling studies, and particularly in a clinical context. First, there are numerous technical problems which make the processing and interpretation of EEG data challenging. EEG—like MEG—is measured on the scalp, and the problem of projecting this 2D-space into the 3D-brain space arises (Michel and Brunet 2019). While multiple solutions exist for this inverse problem, it is unclear which one is the best and under which circumstances (Hassan et al. 2014; Mahjoory et al. 2017; Hedrich et al. 2017). It is important to point out that the goal of source reconstruction is not necessarily to mimic the underlying brain activity, but rather to identify the spatial origins of the signal recorded at the scalp. The question of what constitutes a “source” is still controversial in this context, and the definition depends on the spatial scale (Nunez et al. 2019). Thus, source activity has to be interpreted carefully, taking into account varying degrees of abstraction. Since this is a complex problem involving biophysical mechanisms, we leave this topic aside and recommend the review by Nunez et al. (2019), which addresses the problem of source localization by means of computational models of neural activity on different spatial scales.
EEG data require extensive preprocessing, e.g. removal of artifacts due to movements, eye blinks, etc., but these steps are far from being standardized, and many options exist. The recently started EEG-BIDS effort (Pernet et al. 2019) is a step towards the direction of standardization of EEG data and should facilitate, alongside with the much larger amount of publicly available data, studies that systematically evaluate the impact of preprocessing steps and compare source reconstruction algorithms. As the interest in EEG rises, the need to resolve these issues will trigger larger efforts that will benefit the entire community.
The second obstacle to a more routine usage of computational models in EEG research, which we hope to address in this review, is that such models usually require an understanding of the mathematics involved, if only to be able to choose the model that is useful for the desired application. Both variables and parameters are not always clearly related to quantities which can be measured in a clinical or experimental context, and more generally, models need to be set up in such a way that they meet existing clinical demands or research questions.
The contribution of this paper is threefold. First, this article summarizes computational approaches at different spatial scales in EEG, targeting non-experts readers. To the best of our knowledge, this paper represents the first review on this topic. Second, we will point out several ways in which computational models integrate EEG recordings, by using biologically relevant variables. Third, we discuss the clinical applications of computational models in EEG which have been developed. The field is greatly expanding and contains promising advancements both from research and clinical standpoints. We believe that this overview will make the field accessible for a broad audience, and indicate the next steps required to push modeling of EEG forward.
Electroencephalography
EEG is a non-invasive neuroimaging technique that measures the electrical activity of the brain (Nunez et al. 2006; Schomer and Lopes Da Silva 2012; Biasiucci et al. 2019). EEG recordings have been a driver of research and clinical applications in neuroscience and neurology for nearly a century. EEG relies on the placement of electrodes on the person’s scalp, measuring the postsynaptic potentials of pyramidal neurons (Tivadar and Murray 2019; Lopes da Silva 2013). EEG does not directly measure the action potentials of neurons, though there are some indications of high-frequency oscillations being linked to spiking activity (Telenczuk et al. 2011).
The neurotransmitter release generated by action potentials, whether excitatory or inhibitory, results in local currents at the apical dendrites that in turn lead to current sources and sinks in the extracellular space around the dendritic arbor (i.e. postsynaptic potentials, see Fig. 1, bottom right block).
EEG shares sources with the local field potential (LFP), a low-pass filtered signal of extracellular measurements which represents the summed synaptic activity of local populations of neurons. In the neocortex, pyramidal neurons are generally organized perpendicularly to the cortical surface, with apical dendrites toward the pial surface and axons pointing inferiorly towards the grey-white matter border. This alignment leads to the electrical fields of many neurons being summed up to generate a signal that is measurable at the scalp (Tivadar and Murray 2019).
Importantly, individual neurons of these populations need to be (nearly) synchronously active to be detectable by EEG. When such large-scale synchronization occurs, it manifests in the EEG as oscillations, i.e. sustained sinusoidal activity with a characteristic frequency. The spectral properties of these oscillations—evaluated using the power spectrum—depend on cell types and their connectivity, but they also reflect the “brain state” (neurotransmitters, stimuli, disorders, etc.; see also section “Applications of computational models of EEG”) (Nunez et al. 2006).
As mentioned above, the electrical activity of the brain is recorded by means of electrodes, made of conductive materials, placed at the scalp. The propagation of electrical fields takes place due to the conductive properties of brain and head tissues, a phenomenon known as volume conduction (Kajikawa and Schroeder 2011). The electrodes are connected to an amplifier which boosts the signal. Due to the biophysical nature of what is measured, i.e. a voltage—the difference of potential able to move charges from one site to the other—EEG records the differential measurements between an electrode at a specific position on the scalp and a reference site. Common analyses in EEG are the study of local phenomena such as peaks at specific latencies or scalp sites (event-related potentials, ERPs); or the study of topography, i.e. the shape of the electric field at the scalp, which represents a global brain signature (Murray et al. 2008). EEG is known for its high temporal resolution. The biggest pitfalls of the technique are, on the other hand, the low spatial resolution and signal to noise ratio. A clear and exhaustive walk through these topics as well as an overview of strengths and pitfalls of using EEG is contained in Tivadar and Murray (2019) and for non-experts of the field in Biasiucci et al. (2019).
Despite being a measurement of the scalp activity, EEG can reveal the underlying neurophysiological mechanisms of the brain, and that is what classifies it as brain imaging tool. The estimation of the loci of active sources for the recorded brain activity at the scalp is called source reconstruction (Michel et al. 2004). However, the loci can belong to areas not necessarily below the considered electrode, a pitfall caused by volume conduction. Source reconstruction is a mathematically ill-posed inverse problem, as the solution is not unique. However, the addition of biophysical constraints to the inverse problem allows to retrieve a solution, which has been validated by means of intracranial recordings (Michel and Murray 2012). Having obtained the source activity, one can estimate the functional connectivity between the sources, i.e. the statistical dependencies between brain areas, assumed to indicate their interactions (see also Table 1). This can then be complemented with neuroanatomical/structural connectivity (Table 1), which estimates white matter connections between brain areas.
Computational models stand at the interface between the physiology of neurons at different scales (single neuron, population, macro-scale) and perceptual behavior. EEG would greatly benefit from the integration of in-silico simulations, as computational models could complement both the neurophysiological and behavioral interpretations of EEG recordings. In the following sections, we will discuss different types of computational models, i.e. the different scales at which the neural activity is simulated, how such models can be integrated in the analysis of EEG signals, and how such models have been used in new clinical applications.
Different types of computational models for EEG
A straightforward classification of computational models for EEG can be done based on the different scales of neurophysiological activity they integrate. For instance, we can distinguish three types of models (Fig. 2a):
-
1.
Microscopic models on the level of single cells and micro-circuits;
-
2.
Mesoscopic models on the level of neural masses and neural fields;
-
3.
Macroscopic models taking into account the connectome/white matter.
The integration of computational models has greatly advanced the field of applications of EEG, both for research and clinical purposes.
a Illustration of computational models at the three scales treated here. Microscopic scale: Simple example of two (\(i=1,2\)) leaky integrate-and-fire (LIF) neurons coupled together, a pyramidal neuron making an excitatory synapse to the interneuron, which in turn makes an inhibitory synapse to the pyramidal cell. This minimal circuit implements feedback inhibition, as the pyramidal cell, when activated, will excite the interneuron, which in turn will inhibit it. In the equation, \(V_i\) is the membrane potential of each of the two cells \(i = 1,2\); \(V_L\) is the leak, or resting potential of the cells; R is a constant corresponding to the membrane resistance; \(I_i\) is the synaptic input that each cell receives from the other, and possibly background input; \(\tau\) is the time constant determining how quickly \(V_i\) decays. The model is simulated by setting a firing threshold, at which, when reached, a spike is recorded and \(V_i\) is reset to \(V_L\). Mesoscopic scale: The Wilson–Cowan-model, in which an excitatory (E) and an inhibitory (I) population are coupled together. The mean field equations describe the mean activity of a large number of neurons. \(f_E\) and \(f_I\) are sigmoid transfer functions whose values indicate how many neurons in the population reach firing threshold, and \(h_E\)/\(h_I\) are external inputs like background noise. \(w_{EE}\) and \(w_{II}\) are constants correponding to the strength of self-excitation/inhibition, and \(w_{EI}\) and \(w_{IE}\) the strength of synaptic coupling between populations. Macroscopic scale: In order to simulate long-range interactions between cortical and even subcortical areas, brain network models couple together many mesoscopic (“local”) models using the connection weights defined in the empirical structural connectivity matrix C. The example equation defines the Kuramoto model, in which the phase \(\theta _n\) of each node n is used as a summary of its oscillatory activity around its natural frequency \(\omega\). Each node’s phase depends on the phases of connected nodes p taking into account the time delay \(\tau _{np}\), defined by the distances between nodes n and p. k is a global scaling parameter controlling the strength of internode connections. b Illustration of a typical modeling approach at the macroscopic scale. Activity is simulated for each node using the defined macroscopic model, e.g. the Kuramoto model from panel a, right. The feature of interest is then computed from this activity. Shown here is the functional connectivity, e.g., phase locking values between nodes (Table 1). This can then be compared to the empirical functional connectivity matrix computed in exactly the same way from experimental data, e.g. by correlating the entries of the matrix. The model fit can be determined depending on parameters of the model, e.g. the scaling parameter k or the unit speed, here indicated with “tau”
Computational models for EEG on the level of single cells and microcircuits
The purpose of this level of modeling is to address the origin of the EEG signal by investigating the relationship between its features and electrophysiological mechanisms (Fig. 1, column A) with the tools of computational neuroscience. As detailed above, the EEG signal recorded from the scalp is the result of the spatial integration of the potential fluctuations in the extracellular medium. The EEG signal is mainly caused by the same mechanisms that generate the local field potential (LFP), i.e. it is driven by synaptic activity (Logothetis 2003; Buzsáki et al. 2012) and volume conduction (Kajikawa and Schroeder 2011). From the experimental standpoint, local network activity is usually measured as LFP (mainly in vivo—and rarely in vitro—animal data). By virtue of superposition, fluctuations in the LFP, and EEG more generally, are signatures of correlated neural activity (Pesaran et al. 2018). Cellular and microcircuit modeling are thus aimed at understanding the neurophysiological underpinnings of these correlations and the role played by cell types, connectivity and other properties in shaping the collective activity of neurons.
A primary goal of EEG modeling at the microscopic scale is on the one hand to predict the EEG signal generated by the summation of local dynamics on the microscopic scale and, on the other hand, to reconstruct the microscopic neural activity underlying the observed EEG. The first goal is far from being achieved, and the second is ill-posed due to the number of possible circuit and cellular combinations at the source level leading to similar EEGs. Implicit to these goals is to understand how features of neural circuits, such as the architecture, synapses and cell types, contribute to the generation of electromagnetic fields and their properties in a bottom-up fashion. Despite key insights, many shortcomings limit the interpretability of microcircuit models and the establishment of a one-to-one correspondence with EEG data. For instance, the contribution of spiking activity and correlated cellular fluctuations to LFPs and EEG power spectra remains unclear. Most microcircuit models characterize the net local network activity—used as a proxy for EEG—using the average firing rate or via the mean somatic membrane potential taken amongst populations of cells (of various types). Other studies have used a heuristic approach and approximated the EEG signal as a linear combination of somatic membrane potentials with random coefficients to account for both conduction effects and observational noise (Herrmann et al. 2016; Lefebvre et al. 2017). As such, microcircuit model predictions and experimental data cannot always be compared directly.
Cellular multicompartmental models, which oftentimes take cellular morphology and spatial configuration into consideration, are based on the celebrated Hodgkin-Huxley equations, which describe the temporal evolution of ionic flux across neuronal membranes (see Catterall et al. (2012), for a recent review). Such conductance-based models, which possess explicit and spatially distributed representations for cellular potentials, facilitate the prediction and/or comparison with LFP recordings. In contrast, single compartment models are difficult to interpret: while more abstract single compartment models such as Poisson neurons or integrate-and-fire models (Fig. 2a, left) are often used for their relative tractability and computational efficiency to construct more elaborate microcircuit models, they generally lack the neurophysiological richness to estimate EEG traces. Despite this, several computational advancements in recent years investigated how networks of integrate-and-fire neurons generate LFPs, clarifying the microscopic dynamics reflected in the EEG signal (Mazzoni et al. 2008, 2010, 2011, 2015; Deco et al. 2008; Buehlmann and Deco 2008; Barbieri et al. 2014). Such approaches have been used to understand the formation of correlated activity patterns in the hippocampus (e.g. oscillations), and their associated spectral fingerprints in the LFP (Chatzikalymniou and Skinner 2018). Furthermore, a broad range of works modeled the origin of the local field potential and how it diffuses via volume conduction to generate the EEG signal (Hindriks et al. 2017; Lindén et al. 2011; Mäki-Marttunen et al. 2019; Skaar et al. 2019; Einevoll et al. 2013; Teleńczuk et al. 2017; Bédard and Destexhe 2009).
The key missing element for understanding the link between spiking network activity, LFP, and EEG signal, is the functional and spatial architecture of the networks. In particular, there are two open challenges. The first is to understand how the network connectivity affects the model dynamics that generate the LFP, and the second is to clarify how the spatial arrangement and morphology of neurons affect LFP diffusion (Mazzoni et al. 2015).
From this perspective, models of pyramidal cell dynamics and circuits should guide the interpretation of the EEG signal. For example, Destexhe and colleagues recently addressed the long-debated issue of the relative contribution of inhibitory and excitatory signals to the extracellular signal (Teleńczuk et al. 2019), suggesting that the main source of the EEG signal may stem from inhibitory—rather than excitatory—inputs to pyramidal cells. A recent spiking network model (Saponati et al. 2019) incorporates the modular architecture of the thalamus, in which subnetworks connect to different parts of the cortex (Barardi et al. 2016). This model was used to show how the propagation of activity from the thalamus shapes gamma oscillations in the cortex.
Computational models at the level of single cells and microcircuits have also been instrumental in elucidating the mechanisms underlying multiple EEG phenomena. For instance, such models were used to better understand EEG rhythm changes observed before, during and after anesthesia, using spiking network models (McCarthy et al. 2008; Ching et al. 2012) and/or cortical micro-circuit models (Hutt et al. 2018). Some of these models have been extended to account for the effect of thalamocortical dynamics on EEG oscillations (Ching et al. 2010; Hutt et al. 2018), highlighting the key role played by the thalamus on shaping EEG dynamics. In addition, microcircuit models have been used to understand the EEG response of cortical networks to non-invasive brain stimulation (e.g. TACS, TMS), especially in regard to the interaction between endogenous EEG oscillatory activity and stimulation patterns (Herrmann et al. 2016), in which thalamic interactions were found to play an important role (Lefebvre et al. 2017).
Computational models for EEG on the level of neural masses and neural fields
In this section we discuss models of population dynamics and how they could determine specific features of the electrical activity recorded by EEG (Fig. 1, column B). Mean field models describe the average activity of a large population of neurons by modeling how the population—as a whole—transforms its input currents into an average output firing rate (Fig. 2a, middle; for details on how networks of spiking neurons are reduced to mean field formulations, see Wong and Wang 2006; Deco et al. 2013b; Coombes and Byrne 2019; Byrne et al. 2020). If we consider a population to be a small portion of the cortex containing pyramidal cells, the average activity modelled by the mean field can be understood as the LFP. Two types of models can be distinguished: neural mass models, where variables are a function of time only, and neural field models, where variables are functions of time and space. In this sense, neural field models can be seen as an extension of neural mass models, by taking into account the continuous shape of cortical tissue and the spatial distribution of neurons. These models allow for the description of local lateral inhibition as well as local axonal delays (Hutt et al. 2003; Atay and Hutt 2006). An important application of neural field models is found in phenomenological models of visual hallucinations (Ermentrout and Cowan 1979; Bressloff et al. 2001), and they have been used to model sleep and anaesthesia (Steyn-Ross et al. 1999; Bojak and Liley 2005; Hindriks and van Putten 2012; Hashemi et al. 2015). Future applications may also involve both neural mass and neural field models to describe different cortical structures, similarly to the multiscale approach proposed in Cattani et al. (2016).
The most popular model on this mesoscopic scale was first described by Wilson and Cowan (Wilson and Cowan 1973; Cowan et al. 2016) (Fig. 2a, middle), and all mean field models can be seen as deriving from this form. It consists of an inhibitory and an excitatory population, where usually, for the purpose of EEG, it is assumed that the excitatory population models pyramidal neurons while the inhibitory population takes the role of interneurons. A variant of this model was described in Jansen and Rit (1995) and goes back to the “lumped parameter” model by Lopes Da Silva et al. (1974). It uses three distinct populations, i.e. a population of excitatory interneurons in addition to the two populations already mentioned. The reason this model has been popular in EEG modeling is that it accounts for the observation that inhibitory and excitatory synapses tend to deliver inputs to different parts of the pyramidal cell body (Sotero et al. 2007). In addition, thalamocortical loops are thought to greatly contribute to the generation of oscillations observed in the cortex (Steriade et al. 1993), and an important class of neural field models deals with these loops and their dependency on external stimuli (Robinson et al. 2001b, 2002).
The dynamical behavior of models can be manipulated to simulate different phenomena by varying their parameters. For example, the coupling parameters that determine the strength and speed of feedback-inhibition and feedforward-excitation can be varied (parameters \(w_{IE}\) and \(w_{EI}\) in Fig. 2a, middle), both within and between populations. Also it is possible to modify time constants (which govern the decay of activity in the local populations) or the strength of background noise. Changing these parameters in silico can be interpreted biologically. For example, in Bojak and Liley (2005), the authors describe how a modified neural field model reproduces EEG spectra recorded during anaesthesia. The strength of inputs from the thalamus to the cortical neural populations was varied within a biologically plausible range.
By coupling together more than one model/set of populations, one can start investigating the effect that delays have on neural activity (Jirsa and Haken 1996). In fact, Jansen and Rit (1995) coupled together two neural mass models in order to simulate the effect of interactions between cortical columns on their activity.
Neural mass and neural field models are able to reproduce a range of dynamical behaviors that are observed in EEG, like oscillations in typical EEG frequency bands (David and Friston 2003), phase-amplitude-coupling (Onslow et al. 2014; Sotero 2016), evoked responses (Jansen et al. 1993; Jansen and Rit 1995; David et al. 2005), and power spectra (Lopes Da Silva et al. 1974; Robinson et al. 2001b; David and Friston 2003; Bojak and Liley 2005; Moran et al. 2007). Power spectra are of particular interest in EEG because on the one hand, they can be precisely measured due to the high temporal resolution, and on the other, they can be thought of as a low-dimensional representation of steady-state dynamics. Consequently, much of EEG research focuses on studying shifts in the power spectrum due to task conditions, different cognitive states, or disorders. Of particular interest are linearized versions of these models, which make it possible to estimate the EEG spectrum in an analytical manner (Lopes Da Silva et al. 1974; Robinson et al. 2001b; Liley et al. 2002; Bojak and Liley 2005; Moran et al. 2007; Van Albada et al. 2010). Such solutions are not only more easily interpretable in terms of the impact of varying different parameters and more computationally efficient. They also allow the researcher to quantify the contribution of nonlinearities to the observed power spectra, thus tackling the question of which level of complexity is necessary in computational modeling of EEG.
Often, activity simulated by mean field models is assumed to be related to local field potentials (Liley et al. 2002). However, models are usually set up such that the local field potential derives directly from the mean firing rate. In this way, an important aspect that underlies the EEG signal is neglected, namely, the synchrony (coherence) of the firing within a neural population (as opposed to synchrony between populations, which can be studied using e.g. instantaneous phase differences (Breakspear et al. 2004)). Phenomena such as event-related synchronization and -desynchronization result from a change in this synchrony rather than from a change in firing rate. Recent models (Byrne et al. 2017, 2020) propose therefore a link between the firing rate and the Kuramoto order parameter, which is a measure of how dispersed firing is within a population.
Macroscopic computational models for EEG taking into account the connectome
In this section, we review existing literature on macroscopic computational models that take into account the connectome and discuss their potential to reveal the generative mechanisms of the macroscopic brain activity patterns detected with EEG and MEG (Fig. 1, column C). We will use the term “brain network models” (BNM) in order to clearly distinguish this framework from other approaches to whole-brain modeling (Breakspear 2017), e.g. using neural field models (Jirsa and Haken 1996; Robinson et al. 1997; Coombes et al. 2007) or expansions of the thalamocortical models discussed above (Robinson et al. 2001b; Freyer et al. 2011). We will also leave aside the large body of literature on dynamic causal modeling (DCM) (Kiebel et al. 2008; Pinotsis et al. 2012), as this deserves a more detailed review than the scope of this paper can provide.
Brain network models. In recent years, the interest in the human connectome has experienced a boom, creating the prolific and successful field of “connectomics”. In the framework of connectomics, the brain is conceptualized as a network made up of nodes and edges. Each node represents a brain region, and nodes are coupled together according to a weighted matrix representing the wiring structure of the brain (Fig. 2a, right). This so-called structural connectivity matrix (SC) is derived from white matter fiber bundles which connect distant brain regions (Behrens et al. 2003; Zhang et al. 2010; Hagmann et al. 2008; Sepulcre et al. 2010; Wedeen et al. 2012) and are measured using diffusion weighted magnetic resonance imaging (dMRI) (Table 1). The set of all fiber bundles is called the connectome (Sporns 2011). In practice, brain regions are defined according to an existing brain atlas (for example Desikan et al. (2006); Hagmann et al. (2008); Glasser et al. (2016)), and the weights in the SC matrix are taken as the fiber count (number of streamlines found by a fiber tracking algorithm), fiber density (number of streamlines divided by region size), or, less commonly, some other diffusion imaging-derived quantity, e.g. fractional anisotropy (Wedeen et al. 2008; Iturria-Medina et al. 2008; Essayed et al. 2017). By coupling brain regions together according to the weights in the SC, the activity generated in each region depends also on the activity propagated from other regions along the connections given by the SC. Note that in the previous section, we already mentioned the possibility of coupling local populations. However, in those cases, the coupling is usually determined by a kernel which defines a dependency of the coupling on the geodesic distance between populations.
BNMs are used to study the role of structural connectivity in shaping brain activity patterns. Because this is a complex problem that involves the entire brain, it is important to find a balance between realism and reduction, so that useful predictions can be made. In practice, a common simplification is to assume that all brain regions are largely identical in their dynamical properties (Passingham et al. 2002). This reductionist approach keeps the number of parameters at a manageable level and still allows to investigate how collective phenomena emerge from the realistic connectivity between nodes. In other words, BNMs do not necessarily aim at maximizing the fit to the empirically recorded brain signals. Rather, the goal is to reproduce specific temporal, spatial or spectral features of the empirical data emerging at the macroscopic scale whose underlying mechanisms remain unclear (Fig. 2b).
Choice of local model. In mathematical terms, brain activity is simulated according to a system of coupled differential equations. The activity of each node is described by a mean-field model, such as the ones described in the section “Computational models for EEG on the level of single cells and microcircuits”, and coupling between the mean field models is parametrized by the empirical SC (Fig. 1a, right).
Importantly, the type of mean-field model used at the local level must be selected according to the hypothesis being tested. For example, BNMs have proved to be a powerful tool to elucidate the non-linear link between the brain’s structural wiring and the functional patterns of brain activity captured with resting-state functional magnetic resonance imaging (rsfMRI) (Deco et al. 2013a, 2014a; Honey et al. 2009; Deco et al. 2009; Cabral et al. 2011). However, oscillations in frequency ranges important for M/EEG (2–100 Hz) are often neglected in studies aiming at reproducing correlated fluctuations on the slow time scale of the fMRI signal. Thus, despite the insights gained by BNMs to understand rsfMRI signal dynamics, the same models do not necessarily serve to understand M/EEG signals and vice-versa (Cabral et al. 2017).
In Cabral et al. (2014), the local model employed includes a mechanism for the generation of collective oscillatory signals in order to address oscillatory components of M/EEG. To model brain-wide interactions between local nodes oscillating around a given natural frequency (in this case, 40 Hz, in the gamma frequency range), the Kuramoto model (Kuramoto 2003; Yeung and Strogatz 1999), was extended to incorporate realistic brain connectivity (SC) and time delays (determined by the lengths of the fibers in the SC (see also Finger et al. (2016); Fig. 2a, right). This model shows how, for a specific range of parameters, groups of nodes (communities) can temporarily synchronize at community-specific lower frequencies, obeying to universal rules that govern the behaviour of coupled oscillators with time delays. Thus, the model proposed a mechanism that explains how slow global rhythms in the alpha- and beta-range emerge from interactions of fast local (gamma) oscillations generated by neuronal networks.
In contrast, Deco et al. (2019b) used a mean field model (Wilson and Cowan 1973; Brunel and Wang 2001; Deco and Jirsa 2012; Deco et al. 2014b), which was tuned not to exhibit intrinsic oscillations. Because the brain could thus be considered as being in a noisy, low-activity state, the number of parameters was sufficiently reduced to investigate how activation patterns change over time on different time scales. Time scales including that of M/EEG (ten to several 100 ms) as well as that typical for fMRI (1–3 s) were considered, and the question was asked whether there is a time scale at which brain dynamics are particularly rich. The authors found that both the number of co-activation patterns as well as their dynamics were richest when a time scale of 200 ms was used. Thus, in this case, co-activation patterns were of interest instead of oscillations, and a model suitable for both M/EEG and fMRI was chosen.
Emerging class of harmonics-based models. Although both the described BNMs as well as DCM (dynamic causal modeling) have a long history of success in modeling brain activity patterns, they have high-dimensionality, and usually require local oscillators governed by region-specific or spatially-varying model parameters. While this imbues such models with rich features capable of recreating complex behavior, they are challenging for some clinical applications where a small set of global features might be desired to assess the effect of disease on network activity.
Nunez (1974) presented pioneering modeling work that focuses on the global aspects of brain dynamics, which was continuously developed over the last decades (Nunez 1974, 1989; Nunez and Srinivasan 2006; Nunez et al. 2019). The idea at the basis of these models is that global brain dynamics can be understood as standing and traveling waves constrained by the brain geometry, an idea that remains immensely influential.
In order to take advantage of the low-dimensional properties of such models, some laboratories have recently focused on low-dimensional processes involving diffusion or random walks (Table 1) on the structural graph (Table 1) instead of mean-field models, providing a simpler means of simulating functional connectivity (FC). These simpler models were able to match or exceed the predictive power of complex neural mass models or DCMs in predicting empirical FC (Abdelnour et al. 2014). Higher-order walks on graphs have also been quite successful; typically these methods involve a series expansion of the graph adjacency or Laplacian matrices (Meier et al. 2016; Becker et al. 2018) (Table 1). Not surprisingly, the diffusion and series expansion methods are closely related, and most of these approaches may be interpreted as special cases of each other, as demonstrated elegantly in recent studies (Robinson et al. 2016; Deslauriers-Gauthier et al. 2020; Tewarie et al. 2020).
Whether using graph diffusion or series expansion, these models of spread naturally employ the so-called eigenmodes, or harmonics, of graph adjacency or Laplacian matrix. Hence these methods were generalized to yield spectral graph models whereby e.g. Laplacian harmonics were sufficient to reproduce empirical FC, using only a few eigenmodes (Galán 2008; Atasoy et al. 2016; Abdelnour et al. 2018). The Laplacian matrix in particular has a long history in graph modeling, and its eigenmodes are the orthonormal basis of the network and can thus represent arbitrary patterns on the network (Stewart 1999). Such spectral graph models are computationally attractive due to low-dimensionality and more interpretable analytical solutions. The SC’s Laplacian eigenmodes may be thought of as the substrate on which functional patterns of the brain are established via a process of network transmission (Abdelnour et al. 2018; Atasoy et al. 2016; Robinson et al. 2016; Preti and Van De Ville 2019; Glomb et al. 2020). These models were strikingly successful in replicating canonical functional networks, which are stable large scale circuits made up of functionally distinct brain regions distributed across the cortex that were extracted by clustering a large fMRI dataset (Yeo et al. 2011).
While spectral graph models have demonstrated ability to capture essential steady-state, stationary characteristics of real brain activity, they are limited to modeling passive spread without oscillatory behavior. Hence they may not suitably accommodate a larger repertoire of dynamically-varying microstates or rich power spectra at higher frequencies typically observed on EEG or MEG. Capturing the rich repertoire of brain dynamics would require a full accounting of axonal propagation delays as well as local neural population dynamics within graph models, as previously advocated (Cabral et al. 2011). In O’Connor et al. (2002); O’Connor and Robinson (2004), the authors used neural field models (Robinson et al. 2001a) to derive relationships between wave patterns on the cortical sheet and EEG power spectra. This was later extended to spherical geometries (Robinson et al. 2016; Mukta et al. 2017). Roberts et al. (2019) explored traveling waves on the network derived from the SC, and Tewarie et al. (2019) successfully modeled band-specific MEG resting-state networks with a combination of delayed neural mass models and eigenmodes of the structural network (Tewarie et al. 2019), suggesting delayed interactions in a brain’s network give rise to functional patterns constrained by structural eigenmodes.
Recently another effort was undertaken to characterize wide-band brain activity using graph harmonics in closed form (i.e. requiring no time-domain simulations), a rarity in the field of computational neuroscience (Raj et al. 2020). This “spectral graph model” of brain activity produced realistic power spectra that could successfully predict both the spatial as well as temporal properties of MEG empirical recordings (Raj et al. 2020). Intriguingly, the model has very few (six) parameters, all of which are global and not dependent on local oscillations. This method therefore exemplifies the power of graph methods in reproducing more complex and rich repertoire of brain activity, while keeping to a parsimonious approach that does not require the kinds of high-dimensional and non-linear oscillatory models that have traditionally held sway.
Applications of computational models of EEG
Network oscillations captured through EEG are thought to reflect relevant processes for brain function, namely for cognition, memory, perception, and consciousness (Ward 2003). Indeed, oscillatory activity in EEG signals is found to change with a wide range of tasks and to exhibit characteristic features across states of consciousness. Moreover, alterations in oscillatory activity can be a sign of a brain disorder, with EEG commonly used in research and clinical fields to help diagnosis and treatment (Tatum 2014). It is known that coherence across sufficiently large brain regions is necessary for oscillations to be detectable with EEG. However, the mechanisms generating and orchestrating these oscillations at the mesoscopic and macroscopic levels remain mostly unclear. Following different mechanistic scenarios, physiologically and/or theoretically inspired computational models have been shown to reproduce characteristic features of EEG signals, offering a complementary tool to address healthy and disease brain mechanisms, test new clinical hypotheses, and explore new surgical strategies in silico. This section presents a number of computational works that used mostly large-scale network approaches to explain the changes in brain activity observed across the spectrum of consciousness as well as in neuropsychiatric disorders and epilepsy.
Brain models of consciousness
A variety of models have been employed to elucidate the neurophysiological mechanisms underlying different physiological brain states, such as wakefulness and deep sleep (non-rapid eye movement, NREM) (Robinson et al. 2002; Hill and Tononi 2005; Roberts and Robinson 2012; Cona et al. 2014), and pharmacological conditions, such as anesthesia (Steyn-Ross et al. 1999; Sheeba et al. 2008; Ching et al. 2010; Hutt and Longtin 2010; Liley et al. 2010; Hindriks and van Putten 2012; Ching and Brown 2014; Hashemi et al. 2015). Contrasting the activity detected in quiet wakeful rest with the activity detected in deep sleep and/or anesthesia has been the subject of research of a number of computational models addressing signatures of consciousness captured both with EEG (Esser et al. 2009; Ching et al. 2010) and fMRI (Deco et al. 2018, 2019a). In fact, understanding the mechanisms underlying consciousness might have crucial implications for the study of disorders of consciousness (DoC). DoC refer to a class of clinical conditions that may follow a severe brain injury (hypoxic/ischemic or traumatic brain injury) and include coma, vegetative state or unresponsive wakefulness syndrome (VS/UWS), and minimally conscious state (MCS). Coma has been defined as a state of unresponsiveness characterized by the absence of arousal (patients lie with their eyes closed) and, hence, of awareness. VS/UWS denotes a condition of wakefulness with reflex movements but without behavioural signs of awareness, while patients in MCS show unequivocal signs of interaction with the environment.
The current gold standard for clinical assessment of consciousness relies on the Coma Recovery Scale Revised (Giacino et al. 2004), which scores the ability of patients to behaviourally respond to sensory stimuli or commands. However, behavioral-based clinical diagnoses can lead to misclassification of MCS as VS/UWS because some patients may regain consciousness without recovering their ability to understand, move and communicate (Childs et al. 1993; Andrews et al. 1996; Schnakers et al. 2009). A great effort has been devoted to develop advanced imaging and neurophysiological techniques for assessing covert consciousness and to improve diagnostic and prognostic accuracy (Edlow et al. 2017; Bodart et al. 2017; Stender et al. 2014; Bruno et al. 2011; Owen and Coleman 2008; Stender et al. 2016). A novel neurophysiological approach to unravel the capacity of the brain to sustain consciousness exploits Transcranial Magnetic Stimulation (TMS) in combination with EEG (Rosanova et al. 2018; Casarotto et al. 2016). Specifically, the EEG response evoked by TMS in conscious subjects exhibits complex patterns of activation resulting from preserved cortical interactions. In contrast, when unconscious patients are stimulated with TMS, the evoked-response shows a local pattern of activation, similar to the one observed in healthy controls during NREM sleep and anesthesia.
The perturbational complexity index (PCI) (Casali et al. 2013) is an electroencephalographic-derived measure that quantifies the dynamical complexity of TMS-evoked EEG potentials by means of the Lempel-Ziv compression algorithm, showing high values (low compressibility) for complex chains of activation typical of the awake state, and low values (high compressibility) for stereotypical patterns of activation typical of sleep and anesthesia. PCI has been validated on a benchmark population of 150 conscious and unconscious controls and tested on 81 severely brain-injured patients (Casarotto et al. 2016), showing an unprecedented high sensitivity (94.7%) in discriminating conscious from unconscious states.
Although PCI is commonly used to analyze real TMS-EEG data, it can also be applied to simulated data (Bensaid et al. 2019). This recently published modeling approach investigates the physiological mechanisms underlying the generation of complex or stereotypical TMS-evoked EEG responses. The proposed brain network model, named COALIA in Bensaid et al. (2019), describes local dynamics as neural masses (Wendling et al. 2002) that include populations of pyramidal neurons and three different types of interneurons. Each neural mass describes the local field activity of one of 66 cortical brain regions (Desikan et al. 2006). Neural masses are connected with each other through long-range white matter fibers as described above (section “Macroscopic computational models for EEG taking into account the connectome”). EEG signals are then simulated as neural mass activity. A systematic comparison of the complexity of simulated and real TMS-evoked EEG potentials through PCI suggested that the rhythmically patterned thalamocortical activity, typical of sleep, plays a key role in disrupting the complex patterns of activation evoked by TMS (Bensaid et al. 2019). Indeed, this rhythmical thalamocortical activity results in inhibition within the cortex that prevents information from propagating from one brain region to another, and thus disrupts functional integration, i.e. the ability of the brain to integrate information originating from different brain regions or groups of brain regions (Tononi 1998). Along with functional segregation, i.e., the specialization of a brain region to fulfill a certain function, functional integration is necessary to generate complex time-varying patterns of coordinated cortical activity that are typical of the awake brain, and thought to sustain consciousness and cognitive functions (Tononi et al. 1994; Casali et al. 2013; Lord et al. 2017; Demertzi et al. 2019).
Neuropsychiatric disorders
Disruption of integration and segregation balance, which is fundamental for consciousness as mentioned in the section “Brain models of consciousness”, have been linked also to several neuropsychiatric disorders as a result of altered structural and functional connectivity (Bassett and Bullmore 2009; Fornito et al. 2015; Menon 2011; Deco et al. 2015). Among neuropsychiatric disorders, as reviewed in Lord et al. (2017), Alzheimer’s disease is characterized by a decrease in long-range functional connectivity, directly affecting integration between functional modules of the brain (Stam et al. 2007; Sanz-Arigita et al. 2010). Schizophrenia has been linked to a “subtle randomization” of global functional connectivity, such that the so-called “small-world” character of the network is disrupted (Alexander-Bloch et al. 2010; Lynall et al. 2010); a small-world network is characterized by short path lengths and strong modularity, network properties that are thought to promote information processing in the brain (Bassett and Bullmore 2006) (but see Hilgetag and Goulas (2016)). Loss of integration has also been observed in schizophrenia (Damaraju et al. 2014).
As explained in the section “Macroscopic omputational models for EEG taking into account the connectome”, whole-brain computational models provide insights into how anatomical connections shape and constrain functional connectivity (Honey et al. 2009; Deco et al. 2013a, 2014a). Using BNMs, Cabral and colleagues have shown that the alterations reported in schizophrenia (Lynall et al. 2010) can be explained by a decrease in connectivity between brain areas, occurring either at the local or global level and encompassing either axonal or synaptic mechanisms, hence reinforcing the idea of schizophrenia being the behavioural consequence of a multitude of causes disrupting connectivity between brain areas (Cabral et al. 2012a, b).
However, these models have focused on reproducing fMRI findings and are yet to be extended to address alterations in EEG spectral signatures in schizophrenia, namely increased EEG gamma-band power and decreased alpha power (Uhlhaas and Singer 2013), which, following previous modeling insights (Cabral et al. 2014), may arise from reduced coupling between local gamma-band oscillators. Furthermore, BNMs can be employed to test how clinical interventions may help to re-establish healthy network properties such as the balance between integration and segregation or small-worldness (Deco et al. 2018, 2019a).
Epilepsy
Models have been employed to study pathological alterations of network oscillatory activity related to many diseases, including epilepsy (Wendling 2005; Lytton 2008; Stefanescu et al. 2012; Holt and Netoff 2013). Epilepsy is a complex disease which impacts 1% of the world population and is drug resistant in approximately 30% of cases. Due to its intrinsic complexity, epilepsy research has strongly benefited, and will do so even more in the future, from an in silico environment where hypotheses about brain mechanisms of epileptic seizures can be tested in order to guide strategies for surgical, pharmacological and electrical stimulation techniques.
Focal epilepsy is a prototypical example of a disease that involves both local tissue and network properties. Focal epilepsy occurs when seizures originate in one or multiple sites, so-called epileptogenic zones (EZ), before recruiting close and distal non-epileptogenic areas pertaining to the pathological network. Patients with a history of drug-resistant focal epilepsy are candidates for surgery which targets epileptogenic areas and/or critical nodes presumably involved in the epileptogenic network. Successful outcomes of these procedures critically rely on the ability of clinicians to precisely identify the EZ.
A promising modeling approach aims at studying focal epilepsy through a single-subject virtual brain (Soltesz and Staley 2011; Terry et al. 2012; Hutchings et al. 2015; Proix et al. 2017; Bansal et al. 2018), bringing together the description of how seizures start and end (seizure onset and offset, respectively) at a local level (through neural mass models) (Robinson et al. 2002; Wendling et al. 2002; Lopes da Silva et al. 2003; Breakspear et al. 2006; Jirsa et al. 2014) with individual brain connectivity derived from dMRI data. In this personalized approach, a patient’s brain is virtually reconstructed, such that systematic testing of many surgical scenarios is possible. The individual virtual brain approach provides clinicians with additional information, helping them to identify locations which are responsible for starting or propagating the seizure and whose removal would therefore lead to the patient being seizure-free while avoiding functional side effects of removing brain regions and connections (Proix et al. 2017; Olmi et al. 2019). Finally, in silico approaches involving neurostimulation paradigms provide useful insights about how to prevent seizure onset or to interrupt the propagation of partial seizure to large brain areas (Schiff 2012; Stamoulis 2013; Rich et al. 2020).
Discussion
In this paper we introduced different computational model types and their application to EEG, using a simple classification by spatial scale. Clearly not all models in the literature would necessarily belong to one category, but we believe this taxonomy can provide an entry point for non-experts. The main motivation behind this review was to identify obstacles that stand in the way of applying EEG modeling in both a research and clinical context, and to point out future directions that could remove these obstacles.
We have pointed out several recent efforts that have begun to more closely align models and experimental findings. Such integration of theory and experiment guarantees the use of biologically relevant measures within computational models of EEG, a crucial element if one wishes to use EEG models together with acquired data. For example, recent microcircuit models address the gap between theory and experiment by linking average firing rate—a measure of population activity preferred by the modeling community—and local field potential (LFP)—a measure that is generally thought to be a good proxy of the EEG signal (Saponati et al. 2019); recent mean field models explicitly include the contribution of neural synchronization to the LFP (Byrne et al. 2020), thereby integrating experimental knowledge about how the EEG signal is generated (Lopes da Silva 2013); brain network models explore the contribution of empirically measured connectomes to macroscopic brain activity (Cabral et al. 2014); and applications of computational models already exist that use clinical measures to study e.g. coma (Bensaid et al. 2019), epilepsy (Olmi et al. 2019; Proix et al. 2017), and neuropsychiatric disorders (Spiegler et al. 2016; Kunze et al. 2016). Furthermore, some modeling approaches focus on providing a simple model for large-scale dynamics, making results more interpretable both from a theoretical and clinical standpoint (Abdelnour et al. 2018; Raj et al. 2020).
We have reviewed computational models on three spatial scales (Fig. 2a). Each scale models qualitatively different biological processes which can be measured using distinct recording techniques (Varela et al. 2001) (Fig. 1). While EEG records activity at the macro-scale, mechanisms at each scale have an impact on the EEG signal and should therefore inform its interpretation. Therefore, ideally, scales should be combined to provide a complete picture of neural mechanisms underlying EEG activity, something that started to be explored for example in the simulation platform The Virtual Brain (TVB) (Sanz Leon et al. 2013; Falcon et al. 2016) or in studies showing the theoretical relationship between spiking networks and mean field formulations (Wong and Wang 2006; Deco et al. 2013b; Coombes and Byrne 2019; Byrne et al. 2020). Using models in this hierarchical manner is the only way to disentangle different contributions to the EEG signal without using invasive techniques, i.e., to distinguish neural signals (Michel and Murray 2012; Seeber et al. 2019), volume conduction (Hindriks et al. 2017; Lindén et al. 2011; Mäki-Marttunen et al. 2019; Skaar et al. 2019; Einevoll et al. 2013; Teleńczuk et al. 2017; Bédard and Destexhe 2009), and noise. Furthermore, brain disorders can impact brain structure and function on any scale. Using models on multiple scales is necessary if one wishes to understand how pathological changes manifest in clinically measurable EEG signals. Such an understanding would also allow to use EEG to evaluate clinical interventions that affect the micro- or mesoscale (e.g., drugs).
Models can thus play an important role as a “bridge” that connects different fields. In translational applications, knowledge from basic research can be integrated into a model and the model can be designed in such a way that it is useful for a clinical application. An example for a successful “bridge” is the case of Brain Computer Interfaces. In order to realize multi-scale models, researchers working on animal recordings and researchers focusing on non-invasive recordings in humans have to come together with modeling experts that can incorporate findings from both fields in a model.
As an outlook, EEG modeling could play an important part in future endeavors towards precision medicine, or “personal health”. Individual brain models could be used to integrate different sources of data (EEG, fMRI, ECG, etc.) in a “virtual patient”. This could complement data-driven approaches like connectome fingerprinting [in which individuals are identified using their individual connectome (Finn et al. 2015; Pallarés et al. 2018; Abbas et al. 2020)]. The ultimate goal would be to use this virtual patient to tailor diagnosis and therapies around the needs of the patient (Wium-Andersen et al. 2017), reducing the economical burden and patient discomfort of clinical analyses and hospitalization.
References
Abbas K, Amico E, Svaldi DO, Tipnis U, Duong-Tran DA, Liu M, Rajapandian M, Harezlak J, Ances BM, Goñi J (2020) Geff: graph embedding for functional fingerprinting. Neuroimage 221:117181
Abdelnour F, Voss HU, Raj A (2014) Network diffusion accurately models the relationship between structural and functional brain connectivity networks. Neuroimage 90:335–347
Abdelnour F, Dayan M, Devinsky O, Thesen T, Raj A (2018) Functional brain connectivity is predictable from anatomic network’s laplacian eigen-structure. Neuroimage 172:728–739. https://doi.org/10.1016/j.neuroimage.2018.02.016
Abiri R, Borhani S, Sellers EW, Jiang Y, Zhao X (2019) A comprehensive review of EEG-based brain-computer interface paradigms. J Neural Eng 16(1):011001
Alexander-Bloch AF, Gogtay N, Meunier D, Birn R, Clasen L, Lalonde F, Lenroot R, Giedd J, Bullmore ET (2010) Disrupted modularity and local connectivity of brain functional networks in childhood-onset schizophrenia. Front Syst Neurosci 4:147. https://doi.org/10.3389/fnsys.2010.00147
Andrews K, Murphy L, Munday R, Littlewood C (1996) Misdiagnosis of the vegetative state: retrospective study in a rehabilitation unit. BMJ 313(7048):13–16. https://doi.org/10.1136/bmj.313.7048.13
Atasoy S, Donnelly I, Pearson J (2016) Human brain networks function in connectome-specific harmonic waves. Nat Commun 7(1):10340. https://doi.org/10.1038/ncomms10340
Atay FM, Hutt A (2006) Neural fields with distributed transmission speeds and long-range feedback delays. SIAM J Appl Dyn Syst 5(4):670–698
Bansal K, Nakuci J, Muldoon SF (2018) Personalized brain network models for assessing structure-function relationships. Curr Opin Neurobiol 52:42–47. https://doi.org/10.1016/j.conb.2018.04.014
Barardi A, Garcia-Ojalvo G, Mazzoni A (2016) Transition between functional regimes in an integrate-and-fire network model of the thalamus. PLoS ONE 11(9):e0161934
Barbieri F, Mazzoni A, Logothetis NK, Panzeri S, Brunel N (2014) Stimulus dependence of local field potential spectra: experiment versus theory. J Neurosci 34(44):14589–14605. https://doi.org/10.1523/JNEUROSCI.5365-13.2014
Basser PJ, Mattiello J, LeBihan D (1994) Mr diffusion tensor spectroscopy and imaging. Biophys J 66(1):259–267
Bassett DS, Bullmore E (2006) Small-world brain networks. Neuroscientist 12(6):512–523
Bassett DS, Bullmore ET (2009) Human brain networks in health and disease. Curr Opin Neurol 22(4):340–347. https://doi.org/10.1097/WCO.0b013e32832d93dd
Bassett DS, Sporns O (2017) Network neuroscience. Nat Neurosci 20(3):353–364
Becker CO, Pequito S, Pappas GJ, Miller MB, Grafton ST, Bassett DS, Preciado VM (2018) Spectral mapping of brain functional connectivity from diffusion imaging. Sci Rep 8(1):1–15
Behrens TE, Johansen-Berg H, Woolrich M, Smith S, Wheeler-Kingshott C, Boulby P, Barker G, Sillery E, Sheehan K, Ciccarelli O et al (2003) Non-invasive mapping of connections between human thalamus and cortex using diffusion imaging. Nat Neurosci 6(7):750–757
Bensaid S, Modolo J, Merlet I, Wendling F, Benquet P (2019) COALIA: a computational model of human EEG for consciousness research. Front Syst Neurosci 13:59. https://doi.org/10.3389/fnsys.2019.00059
Biasiucci A, Franceschiello B, Murray MM (2019) Electroencephalography. Curr Biol 29(3):R80–R85
Bodart O, Gosseries O, Wannez S, Thibaut A, Annen J, Boly M, Rosanova M, Casali AG, Casarotto S, Tononi G, Massimini M, Laureys S (2017) Measures of metabolism and complexity in the brain of patients with disorders of consciousness. NeuroImage Clin 14:354–362. https://doi.org/10.1016/j.nicl.2017.02.002
Bojak I, Liley D (2005) Modeling the effects of anesthesia on the electroencephalogram. Phys Rev E 71(4):041902
Breakspear M (2017) Dynamic models of large-scale brain activity. Nat Neurosci 20(3):340
Breakspear M, Williams LM, Stam CJ (2004) A novel method for the topographic analysis of neural activity reveals formation and dissolution of ‘dynamic cell assemblies’. J Comput Neurosci 16(1):49–68
Breakspear M, Roberts JA, Terry JR, Rodrigues S, Mahant N, Robinson PA (2006) A unifying explanation of primary generalized seizures through nonlinear brain modeling and bifurcation analysis. Cereb Cortex 16(9):1296–1313. https://doi.org/10.1093/cercor/bhj072
Bressloff PC, Cowan JD, Golubitsky M, Thomas PJ, Wiener MC (2001) Geometric visual hallucinations, euclidean symmetry and the functional architecture of striate cortex. Philos Trans R Soc Lond, Ser B 356(1407):299–330
Brunel N, Wang XJ (2001) Effects of neuromodulation in a cortical network model of object working memory dominated by recurrent inhibition. J Comput Neurosci 11(1):63–85
Bruno MA, Vanhaudenhuyse A, Thibaut A, Moonen G, Laureys S (2011) From unresponsive wakefulness to minimally conscious PLUS and functional locked-in syndromes: recent advances in our understanding of disorders of consciousness. J Neurol 258(7):1373–1384. https://doi.org/10.1007/s00415-011-6114-x
Buehlmann A, Deco G (2008) The neuronal basis of attention: rate versus synchronization modulation. J Neurosci 28(30):7679–7686
Bulárka S, Gontean A (2016) Brain-computer interface review. In: 2016 12th IEEE International Symposium on Electronics and Telecommunications (ISETC). IEEE, pp 219–222
Buzsáki G, Anastassiou C, Koch C (2012) The origin of extracellular fields and currents—EEG, ECOG, LFP and spikes. Nat Rev Neurosci 13(6):407–420
Byrne A, Brookes MJ, Coombes S (2017) A mean field model for movement induced changes in the beta rhythm. J Comput Neurosci 43(2):143–158
Byrne Á, O’Dea RD, Forrester M, Ross J, Coombes S (2020) Next-generation neural mass and field modeling. J Neurophysiol 123(2):726–742
Bédard C, Destexhe A (2009) Macroscopic models of local field potentials and the apparent 1/f noise in brain activity. Biophys J 96(7):2589–2603
Cabral J, Hugues E, Sporns O, Deco G (2011) Role of local network oscillations in resting-state functional connectivity. Neuroimage 57(1):130–139
Cabral J, Hugues E, Kringelbach ML, Deco G (2012a) Modeling the outcome of structural disconnection on resting-state functional connectivity. Neuroimage 62(3):1342–1353
Cabral J, Kringelbach ML, Deco G (2012b) Functional graph alterations in schizophrenia: a result from a global anatomic decoupling? Pharmacopsychiatry 45(S 01):S57–S64
Cabral J, Luckhoo H, Woolrich M, Joensson M, Mohseni H, Baker A, Kringelbach ML, Deco G (2014) Exploring mechanisms of spontaneous functional connectivity in MEG: how delayed network interactions lead to structured amplitude envelopes of band-pass filtered oscillations. Neuroimage 90:423–435
Cabral J, Kringelbach ML, Deco G (2017) Functional connectivity dynamically evolves on multiple time-scales over a static structural connectome: models and mechanisms. Neuroimage 160:84–96
Casali AG, Gosseries O, Rosanova M, Boly M, Sarasso S, Casali KR, Casarotto S, Bruno MA, Laureys S, Tononi G, Massimini M (2013) A theoretically based index of consciousness independent of sensory processing and behavior. Sci Transl Med 5(198):198ra105. https://doi.org/10.1126/scitranslmed.3006294
Casarotto S, Comanducci A, Rosanova M, Sarasso S, Fecchio M, Napolitani M, Pigorini A, Casali G, Trimarchi PD, Boly M, Gosseries O, Bodart O, Curto F, Landi C, Mariotti M, Devalle G, Laureys S, Tononi G, Massimini M (2016) Stratification of unresponsive patients by an independently validated index of brain complexity. Ann Neurol 80(5):718–729. https://doi.org/10.1002/ana.24779
Cattani A, Solinas S, Canuto C (2016) A hybrid model for the computationally-efficient simulation of the cerebellar granular layer. Front Comput Neurosci 10:30
Catterall WA, Raman IM, Robinson HP, Sejnowski TJ, Paulsen O (2012) The hodgkin-huxley heritage: from channels to circuits. J Neurosci 32(41):14064–14073
Chatzikalymniou AP, Skinner FK (2018) Deciphering the contribution of oriens-lacunosum/moleculare (OLM) cells to intrinsic \(\theta\) rhythms using biophysical local field potential (lfp) models. Eneuro 5(4):e0146
Chiken S, Nambu A (2016) Mechanism of deep brain stimulation: inhibition, excitation, or disruption? Neuroscientist 22(3):313–322
Childs NL, Mercer WN, Childs HW (1993) Accuracy of diagnosis of persistent vegetative state. Neurology 43(8):1465–1467. https://doi.org/10.1212/wnl.43.8.1465
Ching S, Brown EN (2014) Modeling the dynamical effects of anesthesia on brain circuits. Curr Opin Neurobiol 25:116–122. https://doi.org/10.1016/j.conb.2013.12.011
Ching S, Cimenser A, Purdon PL, Brown EN, Kopell NJ (2010) Thalamocortical model for a propofol-induced \(\alpha\)-rhythm associated with loss of consciousness. Proc Natl Acad Sci USA 107(52):22665–22670
Ching S, Purdon PL, Vijayan S, Kopell NJ, Brown EN (2012) A neurophysiological-metabolic model for burst suppression. Proc Natl Acad Sci USA 109(8):3095–3100
Chladni EFF (1802) Die Akustik. Breitkopf & Härtel, Leipzig
Colclough GL, Woolrich MW, Tewarie P, Brookes MJ, Quinn AJ, Smith SM (2016) How reliable are MEG resting-state connectivity metrics? Neuroimage 138:284–293
Cona F, Lacanna M, Ursino M (2014) A thalamo-cortical neural mass model for the simulation of brain rhythms during sleep. J Comput Neurosci 37(1):125–148. https://doi.org/10.1007/s10827-013-0493-1
Coombes S, Byrne Á (2019) Next generation neural mass models. In: Nonlinear dynamics in computational neuroscience. Springer, pp 1–16
Coombes S, Venkov N, Shiau L, Bojak I, Liley DT, Laing CR (2007) Modeling electrocortical activity through improved local approximations of integral neural field equations. Phys Rev E 76(5):051901
Cowan JD, Neuman J, van Drongelen W (2016) Wilson-cowan equations for neocortical dynamics. J Math Neurosci 6(1):1–24
Damaraju E, Allen EA, Belger A, Ford JM, McEwen S, Mathalon DH, Mueller BA, Pearlson GD, Potkin SG, Preda A, Turner JA, Vaidya JG, van Erp TG, Calhoun VD (2014) Dynamic functional connectivity analysis reveals transient states of dysconnectivity in schizophrenia. NeuroImage Clin 5:298–308. https://doi.org/10.1016/j.nicl.2014.07.003
David O, Friston KJ (2003) A neural mass model for MEG/EEG: coupling and neuronal dynamics. Neuroimage 20(3):1743–1755
David O, Harrison L, Friston KJ (2005) Modelling event-related responses in the brain. Neuroimage 25(3):756–770
Deco G, Jirsa VK (2012) Ongoing cortical activity at rest: criticality, multistability, and ghost attractors. J Neurosci 32(10):3366–3375
Deco G, Jirsa VK, Robinson PA, Breakspear M, Friston K (2008) The dynamic brain: from spiking neurons to neural masses and cortical fields. PLoS Comput Biol 4(8):e1000092
Deco G, Jirsa V, McIntosh AR, Sporns O, Kötter R (2009) Key role of coupling, delay, and noise in resting brain fluctuations. Proc Natl Acad Sci 106(25):10302–10307
Deco G, Jirsa VK, McIntosh AR (2013a) Resting brains never rest: computational insights into potential cognitive architectures. Trends Neurosci 36(5):268–274. https://doi.org/10.1016/j.tins.2013.03.001
Deco G, Ponce-Alvarez A, Mantini D, Romani GL, Hagmann P, Corbetta M (2013b) Resting-state functional connectivity emerges from structurally and dynamically shaped slow linear fluctuations. J Neurosci 33(27):11239–11252
Deco G, McIntosh AR, Shen K, Hutchison RM, Menon RS, Everling S, Hagmann P, Jirsa VK (2014a) Identification of optimal structural connectivity using functional connectivity and neural modeling. J Neurosci 34(23):7910–7916. https://doi.org/10.1523/JNEUROSCI.4423-13.2014
Deco G, Ponce-Alvarez A, Hagmann P, Romani GL, Mantini D, Corbetta M (2014b) How local excitation-inhibition ratio impacts the whole brain dynamics. J Neurosci 34(23):7886–7898
Deco G, Tononi G, Boly M, Kringelbach ML (2015) Rethinking segregation and integration: contributions of whole-brain modelling. Nat Rev Neurosci 16(7):430–439. https://doi.org/10.1038/nrn3963
Deco G, Cabral J, Saenger VM, Boly M, Tagliazucchi E, Laufs H, Van Someren E, Jobst B, Stevner A, Kringelbach ML (2018) Perturbation of whole-brain dynamics in silico reveals mechanistic differences between brain states. Neuroimage 169:46–56
Deco G, Cruzat J, Cabral J, Tagliazucchi E, Laufs H, Logothetis NK, Kringelbach ML (2019a) Awakening: predicting external stimulation to force transitions between different brain states. Proc Natl Acad Sci 116(36):18088–18097
Deco G, Cruzat J, Kringelbach ML (2019b) Brain songs framework used for discovering the relevant timescale of the human brain. Nat Commun 10(1):1–13
Demertzi A, Tagliazucchi E, Dehaene S, Deco G, Barttfeld P, Raimondo F, Martial C, Fernández-Espejo D, Rohaut B, Voss HU, Schiff ND, Owen AM, Laureys S, Naccache L, Sitt JD (2019) Human consciousness is supported by dynamic complex patterns of brain signal coordination. Sci Adv 5(2):eaat7603. https://doi.org/10.1126/sciadv.aat7603
Desikan RS, Ségonne F, Fischl B, Quinn BT, Dickerson BC, Blacker D, Buckner RL, Dale AM, Maguire RP, Hyman BT et al (2006) An automated labeling system for subdividing the human cerebral cortex on mri scans into gyral based regions of interest. Neuroimage 31(3):968–980
Deslauriers-Gauthier S, Zucchelli M, Frigo M, Deriche R (2020) A unified framework for multimodal structure-function mapping based on eigenmodes. Med Image Anal 66:101799. https://doi.org/10.1016/j.media.2020.101799
Donetti L, Munoz MA (2004) Detecting network communities: a new systematic and efficient algorithm. J Stat Mech 10:P10012
Edlow BL, Chatelle C, Spencer CA, Chu CJ, Bodien YG, O’Connor KL, Hirschberg RE, Hochberg LR, Giacino JT, Rosenthal ES, Wu O (2017) Early detection of consciousness in patients with acute severe traumatic brain injury. Brain 140(9):2399–2414. https://doi.org/10.1093/brain/awx176
Einevoll GT, Kayser C, Logothetis NK, Panzeri S (2013) Modelling and analysis of local field potentials for studying the function of cortical circuits. Nat Rev Neurosci 14(11):770–785
Ermentrout GB, Cowan JD (1979) A mathematical theory of visual hallucination patterns. Biol Cybern 34(3):137–150
Essayed WI, Zhang F, Unadkat P, Cosgrove GR, Golby AJ, O’Donnell LJ (2017) White matter tractography for neurosurgical planning: a topography-based review of the current state of the art. NeuroImage 15:659–672
Esser SK, Hill S, Tononi G (2009) Breakdown of effective connectivity during slow wave sleep: investigating the mechanism underlying a cortical gate using large-scale modeling. J Neurophysiol 102(4):2096–111
Falcon MI, Jirsa V, Solodkin A (2016) A new neuroinformatics approach to personalized medicine in neurology: the virtual brain. Curr Opin Neurol 29(4):429
Feigin VL, Nichols E, Alam T, Bannick MS, Beghi E, Blake N, Culpepper WJ, Dorsey ER, Elbaz A, Ellenbogen RG et al (2019) Global, regional, and national burden of neurological disorders, 1990–2016: a systematic analysis for the global burden of disease study 2016. Lancet Neurol 18(5):459–480
Finger H, Bönstrup M, Cheng B, Messé A, Hilgetag C, Thomalla G, Gerloff C, König P (2016) Modeling of large-scale functional brain networks based on structural connectivity from DTI: comparison with EEG derived phase coupling networks and evaluation of alternative methods along the modeling path. PLoS Comput Biol 12(8):e1005025
Finn ES, Shen X, Scheinost D, Rosenberg MD, Huang J, Chun MM, Papademetris X, Constable RT (2015) Functional connectome fingerprinting: identifying individuals using patterns of brain connectivity. Nat Neurosci 18(11):1664–1671
Fornito A, Zalesky A, Breakspear M (2015) The connectomics of brain disorders. Nat Rev Neurosci 16(3):159–172. https://doi.org/10.1038/nrn3901
Fouad MM, Amin KM, El-Bendary N, Hassanien AE (2015) Brain computer interface: a review. In: Brain-computer interfaces. Springer, pp 3–30
Franceschiello B, Sarti A, Citti G (2018) A neuromathematical model for geometrical optical illusions. J Math Imaging Vis 60(1):94–108
Franceschiello B, Mashtakov A, Citti G, Sarti A (2019) Geometrical optical illusion via sub-riemannian geodesics in the roto-translation group. Differ Geom Appl 65:55–77
Freyer F, Roberts JA, Becker R, Robinson PA, Ritter P, Breakspear M (2011) Biophysical mechanisms of multistability in resting-state cortical rhythms. J Neurosci 31(17):6353–6361
Friston KJ (2011) Functional and effective connectivity: a review. Brain connectivity 1(1):13–36
Galán RF (2008) On how network architecture determines the dominant patterns of spontaneous neural activity. PLoS ONE 3(5):e2148
Giacino JT, Kalmar K, Whyte J (2004) The JFK coma recovery scale-revised: measurement characteristics and diagnostic utility. Arch Phys Med Rehabil 85(12):2020–2029
Glasser MF, Coalson TS, Robinson EC, Hacker CD, Harwell J, Yacoub E, Ugurbil K, Andersson J, Beckmann CF, Jenkinson M et al (2016) A multi-modal parcellation of human cerebral cortex. Nature 536(7615):171–178
Glomb K, Queralt JR, Pascucci D, Defferrard M, Tourbier S, Carboni M, Rubega M, Vulliemoz S, Plomp G, Hagmann P (2020) Connectome spectral analysis to track EEG task dynamics on a subsecond scale. NeuroImage 221:117137
Hagmann P, Cammoun L, Gigandet X, Meuli R, Honey CJ, Wedeen VJ, Sporns O (2008) Mapping the structural core of human cerebral cortex. PLoS Biol 6(7):e159
Hashemi M, Hutt A, Sleigh J (2015) How the cortico-thalamic feedback affects the EEG power spectrum over frontal and occipital regions during propofol-induced sedation. J Comput Neurosci 39(2):155–179
Hassan M, Dufor O, Merlet I, Berrou C, Wendling F (2014) EEG source connectivity analysis: from dense array recordings to brain networks. PLoS ONE 9(8):e105041
Hedrich T, Pellegrino G, Kobayashi E, Lina JM, Grova C (2017) Comparison of the spatial resolution of source imaging techniques in high-density EEG and MEG. Neuroimage 157:531–544
Herrmann CS, Murray MM, Ionta S, Hutt A, Lefebvre J (2016) Shaping intrinsic neural oscillations with periodic stimulation. J Neurosci 36(19):5328–5337
Hilgetag CC, Goulas A (2016) Is the brain really a small-world network? Brain Struct Funct 221(4):2361–2366
Hill S, Tononi G (2005) Modeling sleep and wakefulness in the thalamocortical system. J Neurophysiol 93(3):1671–1698. https://doi.org/10.1152/jn.00915.2004
Hindriks R, van Putten MJ (2012) Meanfield modeling of propofol-induced changes in spontaneous EEG rhythms. Neuroimage 60(4):2323–2334
Hindriks R, Schmiedt J, Arsiwalla XD, Peter A, Verschure PF, Fries P, Schmid MC, Deco G (2017) Linear distributed source modeling of local field potentials recorded with intra-cortical electrode arrays. PLoS ONE 12(12):e0187490
Holt AB, Netoff TI (2013) Computational modeling of epilepsy for an experimental neurologist. Exp Neurol 244:75–86. https://doi.org/10.1016/j.expneurol.2012.05.003
Honey CJ, Sporns O, Cammoun L, Gigandet X, Thiran JP, Meuli R, Hagmann P (2009) Predicting human resting-state functional connectivity from structural connectivity. Proc Natl Acad Sci USA 106(6):2035–2040. https://doi.org/10.1073/pnas.0811168106
Hutchings F, Han CE, Keller SS, Weber B, Taylor PN, Kaiser M (2015) Predicting surgery targets in temporal lobe epilepsy through structural connectome based simulations. PLoS Comput Biol 11(12):e1004642. https://doi.org/10.1371/journal.pcbi.1004642
Hutt A, Longtin A (2010) Effects of the anesthetic agent propofol on neural populations. Cogn Neurodyn 4(1):37–59. https://doi.org/10.1007/s11571-009-9092-2
Hutt A, Bestehorn M, Wennekers T (2003) Pattern formation in intracortical neuronal fields. Network 14(2):351–368
Hutt A, Lefebvre J, Hight D, Sleigh J (2018) Suppression of underlying neuronal fluctuations mediates EEG slowing during general anaesthesia. Neuroimage 179:414–428
Iturria-Medina Y, Sotero RC, Canales-Rodríguez EJ, Alemán-Gómez Y, Melie-García L (2008) Studying the human brain anatomical network via diffusion-weighted mri and graph theory. Neuroimage 40(3):1064–1076
Jansen BH, Rit VG (1995) Electroencephalogram and visual evoked potential generation in a mathematical model of coupled cortical columns. Biol Cybern 73(4):357–366
Jansen BH, Zouridakis G, Brandt ME (1993) A neurophysiologically-based mathematical model of flash visual evoked potentials. Biol Cybern 68(3):275–283
Jeurissen B, Descoteaux M, Mori S, Leemans A (2019) Diffusion mri fiber tractography of the brain. NMR Biomed 32(4):e3785
Jirsa VK, Haken H (1996) Field theory of electromagnetic brain activity. Phys Rev Lett 77(5):960
Jirsa VK, Stacey WC, Quilichini PP, Ivanov AI, Bernard C (2014) On the nature of seizure dynamics. Brain 137(Pt 8):2210–2230. https://doi.org/10.1093/brain/awu133
Jones DK (2010) Challenges and limitations of quantifying brain connectivity in vivo with diffusion MRI. Imaging Med 2(3):341–355
Kajikawa Y, Schroeder CE (2011) How local is the local field potential? Neuron 72(5):847–858
Kiebel SJ, Garrido MI, Moran RJ, Friston KJ (2008) Dynamic causal modelling for EEG and MEG. Cogn Neurodyn 2(2):121
Kunze T, Hunold A, Haueisen J, Jirsa V, Spiegler A (2016) Transcranial direct current stimulation changes resting state functional connectivity: a large-scale brain network modeling study. Neuroimage 140:174–187. https://doi.org/10.1016/j.neuroimage.2016.02.015
Kuramoto Y (2003) Chemical oscillations, waves, and turbulence. Courier Corporation, North Chelmsford
Lefebvre J, Hutt A, Frohlich F (2017) Stochastic resonance mediates the state-dependent effect of periodic stimulation on cortical alpha oscillations. Elife 6:e32054
Liley DT, Cadusch PJ, Dafilis MP (2002) A spatially continuous mean field theory of electrocortical activity. Network 13(1):67–113
Liley DTJ, Sinclair NC, Lipping T, Heyse B, Vereecke HEM, Struys MMRF (2010) Propofol and remifentanil differentially modulate frontal electroencephalographic activity. Anesthesiology 113(2):292–304. https://doi.org/10.1097/ALN.0b013e3181e3d8a6
Lindén H, Tetzlaff T, Potjans TC, Pettersen KH, Grün S, Diesmann M, Einevoll GT (2011) Modeling the spatial reach of the LFP. Neuron 72(5):859–872
Logothetis NK (2003) The underpinnings of the bold functional magnetic resonance imaging signal. J Neurosci 23(10):3963–3971. https://doi.org/10.1523/JNEUROSCI.23-10-03963.2003
Lopes da Silva F (2013) EEG and MEG: relevance to neuroscience. Neuron 80(5):1112–1128
Lopes Da Silva F, Hoeks A, Smits H, Zetterberg L (1974) Model of brain rhythmic activity. Kybernetik 15(1):27–37
Lopes da Silva F, Blanes W, Kalitzin SN, Parra J, Suffczynski P, Velis DN (2003) Epilepsies as dynamical diseases of brain systems: basic models of the transition between normal and epileptic activity. Epilepsia 44(Suppl 12):72–83. https://doi.org/10.1111/j.0013-9580.2003.12005.x
Lord LD, Stevner AB, Deco G, Kringelbach ML (2017) Kringelbach ML (2017) Understanding principles of integration and segregation using whole-brain computational connectomics: implications for neuropsychiatric disorders. Philos Trans Ser A 375:20160283. https://doi.org/10.1098/rsta.2016.0283
Lynall ME, Bassett DS, Kerwin R, McKenna PJ, Kitzbichler M, Muller U, Bullmore E (2010) Functional connectivity and brain networks in schizophrenia. J Neurosci 30(28):9477–9487. https://doi.org/10.1523/JNEUROSCI.0333-10.2010
Lytton WW (2008) Computer modelling of epilepsy. Nat Rev Neurosci 9(8):626–637. https://doi.org/10.1038/nrn2416
Mahjoory K, Nikulin VV, Botrel L, Linkenkaer-Hansen K, Fato MM, Haufe S (2017) Consistency of EEG source localization and connectivity estimates. Neuroimage 152:590–601
Mäki-Marttunen T, Kaufmann T, Elvsåshagen T, Devor A, Djurovic S, Westlye LT, Linne ML, Rietschel M, Schubert D, Borgwardt S et al (2019) Biophysical psychiatry—how computational neuroscience can help to understand the complex mechanisms of mental disorders. Front Psychiatry 10:534
Mazzoni A, Panzeri S, Logothetis NK, Brunel N (2008) Encoding of naturalistic stimuli by local field potential spectra in networks of excitatory and inhibitory neurons. PLoS Comput Biol 4(12):e1000239
Mazzoni A, Whittingstall K, Brunel N, Logothetis NK, Panzeri S (2010) Understanding the relationships between spike rate and delta/gamma frequency bands of LFPS and EEGs using a local cortical network model. Neuroimage 52(3):956–972. https://doi.org/10.1016/j.neuroimage.2009.12.040
Mazzoni A, Brunel N, Cavallari S, Logothetis NK, Panzeri S (2011) Cortical dynamics during naturalistic sensory stimulations: experiments and models. J Physiol 105(1):2–15. https://doi.org/10.1016/j.jphysparis.2011.07.014 computational Neuroscience: Neurocomp 2010
Mazzoni A, Linden H, Cuntz H, Lansner A, Panzeri S, Einevoll GT (2015) Computing the local field potential (LFP) from integrate-and-fire network models. PLoS Comput Biol 11(12):e1004584
McGregor MM, Nelson AB (2019) Circuit mechanisms of parkinson’s disease. Neuron 101(6):1042–1056
Meier J, Tewarie P, Hillebrand A, Douw L, van Dijk BW, Stufflebeam SM, Van Mieghem P (2016) A mapping between structural and functional brain networks. Brain Connectivity 6(4):298–311. https://doi.org/10.1089/brain.2015.0408
Menon V (2011) Large-scale brain networks and psychopathology: a unifying triple network model. Trends Cogn Sci 15(10):483–506. https://doi.org/10.1016/j.tics.2011.08.003
Michel CM, Brunet D (2019) EEG source imaging: a practical review of the analysis steps. Front Neurol 10:325
Michel CM, Murray MM (2012) Towards the utilization of EEG as a brain imaging tool. Neuroimage 61(2):371–385
Michel CM, Murray MM, Lantz G, Gonzalez S, Spinelli L, de Peralta RG (2004) EEG source imaging. Clin Neurophysiol 115(10):2195–2222
McCarthy Michelle M, Emery NK, Brown N (2008) Potential network mechanisms mediating electroencephalographic beta rhythm changes during propofol-induced paradoxical excitation. J Neurosci 28(50):13488–13504
Moran RJ, Kiebel SJ, Stephan KE, Reilly R, Daunizeau J, Friston KJ (2007) A neural mass model of spectral responses in electrophysiology. Neuroimage 37(3):706–720
Mukta K, MacLaurin J, Robinson P (2017) Theory of corticothalamic brain activity in a spherical geometry: spectra, coherence, and correlation. Phys Rev E 96(5):052410
Murray MM, Brunet D, Michel CM (2008) Topographic erp analyses: a step-by-step tutorial review. Brain Topogr 20(4):249–264
Nunez PL (1974) The brain wave equation: a model for the EEG. Math Biosci 21(3–4):279–297
Nunez PL (1989) Generation of human EEG by a combination of long and short range neocortical interactions. Brain Topogr 1(3):199–215
Nunez PL, Srinivasan R (2006) A theoretical basis for standing and traveling brain waves measured with human EEG with implications for an integrated consciousness. Clin Neurophysiol 117(11):2424–2435
Nunez PL, Srinivasan R (2006) Electric fields of the brain: the neurophysics of EEG. Oxford University Press, Oxford
Nunez PL, Nunez MD, Srinivasan R (2019) Multi-scale neural sources of EEG: genuine, equivalent, and representative. a tutorial review. Brain Topogr 32(2):193–214
Olmi S, Petkoski S, Guye M, Bartolomei F, Jirsa V (2019) Controlling seizure propagation in large-scale brain networks. PLoS Comput Biol 15(2):e1006805. https://doi.org/10.1371/journal.pcbi.1006805
Onslow AC, Jones MW, Bogacz R (2014) A canonical circuit for generating phase-amplitude coupling. PLoS ONE 9(8):e102591
Owen AM, Coleman MR (2008) Detecting awareness in the vegetative state. Ann N Y Acad Sci 1129:130–138. https://doi.org/10.1196/annals.1417.018
O’Connor S, Robinson P (2004) Spatially uniform and nonuniform analyses of electroencephalographic dynamics, with application to the topography of the alpha rhythm. Phys Rev E 70(1):011911
O’Connor S, Robinson P, Chiang A (2002) Wave-number spectrum of electroencephalographic signals. Phys Rev E 66(6):061905
Pallarés V, Insabato A, Sanjuán A, Kühn S, Mantini D, Deco G, Gilson M (2018) Extracting orthogonal subject-and condition-specific signatures from fMRI data using whole-brain effective connectivity. Neuroimage 178:238–254
Passingham RE, Stephan KE, Kötter R (2002) The anatomical basis of functional localization in the cortex. Nat Rev Neurosci 3(8):606–616
Pernet CR, Appelhoff S, Gorgolewski KJ, Flandin G, Phillips C, Delorme A, Oostenveld R (2019) EEG-BIDS, an extension to the brain imaging data structure for electroencephalography. Sci Data 6(1):1–5
Pesaran B, Vinck M, Einevoll GT, Sirota A, Fries P, Siegel M, Truccolo W, Schroeder CE, Srinivasan R (2018) Investigating large-scale brain dynamics using field potential recordings: analysis and interpretation. Nat Neurosci 21(7):903–919
Pinotsis DA, Moran RJ, Friston KJ (2012) Dynamic causal modeling with neural fields. Neuroimage 59(2):1261–1274
Preti MG, Van De Ville D (2019) Decoupling of brain function from structure reveals regional behavioral specialization in humans. Nat Commun 10(1):1–7. https://doi.org/10.1038/s41467-019-12765-7
Proix T, Bartolomei F, Guye M, Jirsa VK (2017) Individual brain structure and modelling predict seizure propagation. Brain 140(3):641–654. https://doi.org/10.1093/brain/awx004 publisher: Oxford Academic
Raj A, Cai C, Xie X, Palacios E, Owen J, Mukherjee P, Nagarajan S (2020) Spectral graph theory of brain oscillations. Hum Brain Mapp. https://doi.org/10.1002/hbm.24991
Rich S, Hutt A, Skinner FK, Valiante TA, Lefebvre J (2020) Neurostimulation stabilizes spiking neural networks by disrupting seizure-like oscillatory transitions. Sci Rep 10(1):15408
Roberts JA, Robinson PA (2012) Corticothalamic dynamics: structure of parameter space, spectra, instabilities, and reduced model. Phys Rev E 85(1 Pt 1):011910. https://doi.org/10.1103/PhysRevE.85.011910
Roberts JA, Gollo LL, Abeysuriya RG, Roberts G, Mitchell PB, Woolrich MW, Breakspear M (2019) Metastable brain waves. Nat Commun 10(1):1–17
Robinson P, Loxley P, O’connor S, Rennie C (2001a) Modal analysis of corticothalamic dynamics, electroencephalographic spectra, and evoked potentials. Phys Rev E 63(4):041909
Robinson P, Rennie C, Wright J, Bahramali H, Gordon E, Rowe D (2001b) Prediction of electroencephalographic spectra from neurophysiology. Phys Rev E 63(2):021903
Robinson PA, Rennie CJ, Wright JJ (1997) Propagation and stability of waves of electrical activity in the cerebral cortex. Phys Rev E 56(1):826
Robinson PA, Rennie CJ, Rowe DL (2002) Dynamics of large-scale brain activity in normal arousal states and epileptic seizures. Phys Rev E 65(4 Pt 1):041924. https://doi.org/10.1103/PhysRevE.65.041924
Robinson PA, Zhao X, Aquino KM, Griffiths J, Sarkar S, Mehta-Pandejee G (2016) Eigenmodes of brain activity: neural field theory predictions and comparison with experiment. Neuroimage 142:79–98
Rosanova M, Fecchio M, Casarotto S, Sarasso S, Casali AG, Pigorini A, Comanducci A, Seregni F, Devalle G, Citerio G, Bodart O, Boly M, Gosseries O, Laureys S, Massimini M (2018) Sleep-like cortical OFF-periods disrupt causality and complexity in the brain of unresponsive wakefulness syndrome patients. Nat Commun 9(1):4427. https://doi.org/10.1038/s41467-018-06871-1
Rubinov M, Sporns O (2010) Complex network measures of brain connectivity: uses and interpretations. Neuroimage 52(3):1059–1069
Sanz-Arigita EJ, Schoonheim MM, Damoiseaux JS, Rombouts SARB, Maris E, Barkhof F, Scheltens P, Stam CJ (2010) Loss of ‘small-world’ networks in Alzheimer’s disease: graph analysis of fMRI resting-state functional connectivity. PLoS ONE 5(11):e13788. https://doi.org/10.1371/journal.pone.0013788
Sanz Leon P, Knock SA, Woodman MM, Domide L, Mersmann J, McIntosh AR, Jirsa V (2013) The virtual brain: a simulator of primate brain network dynamics. Front Neuroinform 7:10
Saponati M, Garcia-Ojalvo J, Cataldo E, Mazzoni A (2019) Integrate-and-fire network model of activity propagation from thalamus to cortex. Biosystems 183:103978
Schiff SJ (2012) Neural control engineering. In: Sejnowski TJ, Poggio TA (eds) Computational neuroscience. MIT Press, Cambridge
Schnakers C, Vanhaudenhuyse A, Giacino J, Ventura M, Boly M, Majerus S, Moonen G, Laureys S (2009) Diagnostic accuracy of the vegetative and minimally conscious state: clinical consensus versus standardized neurobehavioral assessment. BMC Neurol 9:35. https://doi.org/10.1186/1471-2377-9-35
Schomer DL, Lopes Da Silva F (2012) Niedermeyer’s electroencephalography: basic principles, clinical applications, and related fields. Lippincott Williams & Wilkins, Philadelphia
Seeber M, Cantonas LM, Hoevels M, Sesia T, Visser-Vandewalle V, Michel CM (2019) Subcortical electrophysiological activity is detectable with high-density EEG source imaging. Nat Commun 10(1):1–7
Sepulcre J, Liu H, Talukdar T, Martincorena I, Yeo BT, Buckner RL (2010) The organization of local and distant functional connectivity in the human brain. PLoS Comput Biol 6(6):e1000808
Sheeba JH, Stefanovska A, McClintock PVE (2008) Neuronal synchrony during anesthesia: a thalamocortical model. Biophys J 95(6):2722–2727. https://doi.org/10.1529/biophysj.108.134635
Skaar JEW, Stasik AJ, Hagen E, Ness TV, Einevoll GT (2019) Estimation of neural network model parameters from local field potentials (LFPS). bioRxiv 564765
Soltesz I, Staley K (2011) Computational neuroscience in epilepsy. Academic Press, google-Books-ID: ytc3hT3TMCAC
Sotero RC (2016) Topology, cross-frequency, and same-frequency band interactions shape the generation of phase-amplitude coupling in a neural mass model of a cortical column. PLoS Comput Biol 12(11):e1005180
Sotero RC, Trujillo-Barreto NJ, Iturria-Medina Y, Carbonell F, Jimenez JC (2007) Realistically coupled neural mass models can generate EEG rhythms. Neural Comput 19(2):478–512
Spiegler A, Hansen ECA, Bernard C, McIntosh AR, Jirsa VK (2016) Selective activation of resting-state networks following focal stimulation in a connectome-based network model of the human brain. eNeuro. https://doi.org/10.1523/ENEURO.0068-16.2016
Sporns O (2011) The human connectome: a complex network. Ann N Y Acad Sci 1224(1):109–125
Sporns O (2018) Graph theory methods: applications in brain networks. Dialogues Clin Neurosci 20(2):111
Stam CJ, Jones BF, Nolte G, Breakspear M, Scheltens P (2007) Small-world networks and functional connectivity in Alzheimer’s disease. Cereb Cortex 17(1):92–99. https://doi.org/10.1093/cercor/bhj127
Stamoulis CCB, Stamoulis C, Chang BS (2013) Modeling noninvasive neurostimulation in epilepsy as stochastic interference in brain networks. IEEE Trans Neural Syst Rehabil Eng 21(3):354–363
Steel Z, Marnane C, Iranpour C, Chey T, Jackson JW, Patel V, Silove D (2014) The global prevalence of common mental disorders: a systematic review and meta-analysis 1980–2013. Int J Epidemiol 43(2):476–493
Stefanescu RA, Shivakeshavan RG, Talathi SS (2012) Computational models of epilepsy. Seizure 21(10):748–759. https://doi.org/10.1016/j.seizure.2012.08.012
Stender J, Gosseries O, Bruno MA, Charland-Verville V, Vanhaudenhuyse A, Demertzi A, Chatelle C, Thonnard M, Thibaut A, Heine L, Soddu A, Boly M, Schnakers C, Gjedde A, Laureys S (2014) Diagnostic precision of PET imaging and functional MRI in disorders of consciousness: a clinical validation study. Lancet 384(9942):514–522. https://doi.org/10.1016/S0140-6736(14)60042-8
Stender J, Mortensen KN, Thibaut A, Darkner S, Laureys S, Gjedde A, Kupers R (2016) The minimal energetic requirement of sustained awareness after brain injury. Curr Biol 26(11):1494–1499. https://doi.org/10.1016/j.cub.2016.04.024
Steriade M, McCormick DA, Sejnowski TJ (1993) Thalamocortical oscillations in the sleeping and aroused brain. Science 262(5134):679–685
Stewart I (1999) Holes and hot spots. Nature 401(6756):863–865. https://doi.org/10.1038/44730
Steyn-Ross ML, Steyn-Ross DA, Sleigh JW, Liley D (1999) Theoretical electroencephalogram stationary spectrum for a white-noise-driven cortex: evidence for a general anesthetic-induced phase transition. Phys Rev E 60(6):7299
Tatum WO (2014) Handbook of EEG interpretation, nd edn. Demos Medical Publishing, google-Books-ID: BLsiAwAAQBAJ
Teleńczuk B, Baker SN, Herz AV, Curio G (2011) High-frequency EEG covaries with spike burst patterns detected in cortical neurons. J Neurophysiol 105(6):2951–2959
Teleńczuk B, Dehghani N, Le Van Quyen M, Cash SS, Halgren E, Hatsopoulos NG, Destexhe A (2017) Local field potentials primarily reflect inhibitory neuron activity in human and monkey cortex. Sci Rep 7:40211
Teleńczuk M, Teleńczuk B, Destexhe A (2019) Modeling unitary fields and the single-neuron contribution to local field potentials in the hippocampus. BioRxiv 602953
Terry JR, Benjamin O, Richardson MP (2012) Seizure generation: the role of nodes and networks. Epilepsia 53(9):e166–169. https://doi.org/10.1111/j.1528-1167.2012.03560.x
Tewarie P, Abeysuriya R, Byrne Á, O’Neill GC, Sotiropoulos SN, Brookes MJ, Coombes S (2019) How do spatially distinct frequency specific MEG networks emerge from one underlying structural connectome? The role of the structural eigenmodes. Neuroimage 186:211–220
Tewarie P, Prasse B, Meier J, Santos F, Douw L, Schoonheim M, Stam C, Van Mieghem VanP, Hillebrand A (2020) Mapping functional brain networks from the structural connectome: relating the series expansion and eigenmode approaches. Neuroimage 216:116805. https://doi.org/10.1016/j.neuroimage.2020.116805
Tivadar RI, Murray MM (2019) A primer on electroencephalography and event-related potentials for organizational neuroscience. Organizational Res Methods 22(1):69–94
Tononi G (1998) Consciousness and complexity. Science 282(5395):1846–1851. https://doi.org/10.1126/science.282.5395.1846
Tononi G, Sporns O, Edelman GM (1994) A measure for brain complexity: relating functional segregation and integration in the nervous system. Proc Natl Acad Sci 91(11):5033–5037
Uhlhaas PJ, Singer W (2013) High-frequency oscillations and the neurobiology of schizophrenia. Dialogues Clin Neurosci 15(3):301
Van Albada S, Kerr C, Chiang A, Rennie C, Robinson P (2010) Neurophysiological changes with age probed by inverse modeling of EEG spectra. Clin Neurophysiol 121(1):21–38
Varela F, Lachaux JP, Rodriguez E, Martinerie J (2001) The brainweb: phase synchronization and large-scale integration. Nat Rev Neurosci 2(4):229
Vigo D, Thornicroft G, Atun R (2016) Estimating the true global burden of mental illness. Lancet Psychiatry 3(2):171–178
Ward LM (2003) Synchronous neural oscillations and cognitive processes. Trends Cogn Sci 7(12):553–559. https://doi.org/10.1016/j.tics.2003.10.012
Wedeen VJ, Wang R, Schmahmann JD, Benner T, Tseng WYI, Dai G, Pandya D, Hagmann P, D’Arceuil H, de Crespigny AJ (2008) Diffusion spectrum magnetic resonance imaging (DSI) tractography of crossing fibers. Neuroimage 41(4):1267–1277
Wedeen VJ, Rosene DL, Wang R, Dai G, Mortazavi F, Hagmann P, Kaas JH, Tseng WYI (2012) The geometric structure of the brain fiber pathways. Science 335(6076):1628–1634
Wendling F (2005) Neurocomputational models in the study of epileptic phenomena. J Clin Neurophysiol 22(5):285–287
Wendling F, Bartolomei F, Bellanger JJ, Chauvel P (2002) Epileptic fast activity can be explained by a model of impaired GABAergic dendritic inhibition. Eur J Neurosci 15(9):1499–1508. https://doi.org/10.1046/j.1460-9568.2002.01985.x
Wilson HR, Cowan JD (1973) A mathematical theory of the functional dynamics of cortical and thalamic nervous tissue. Kybernetik 13(2):55–80
Wium-Andersen IK, Vinberg M, Kessing LV, McIntyre RS (2017) Personalized medicine in psychiatry. Nord J Psychiatry 71(1):12–19
Wolpaw JR, Birbaumer N, Heetderks WJ, McFarland DJ, Peckham PH, Schalk G, Donchin E, Quatrano LA, Robinson CJ, Vaughan TM (2000) Brain-computer interface technology: a review of the first international meeting. IEEE Trans Rehabil Eng 8(2):164–173
Wong KF, Wang XJ (2006) A recurrent network mechanism of time integration in perceptual decisions. J Neurosci 26(4):1314–1328
Yeo BTT, Krienen FM, Sepulcre J, Sabuncu MR, Lashkari D, Hollinshead M, Roffman JL, Smoller JW, Zöllei L, Polimeni JR, Fischl B, Liu H, Buckner RL (2011) The organization of the human cerebral cortex estimated by intrinsic functional connectivity. J Neurophysiol 106(3):1125–65. https://doi.org/10.1152/jn.00338.2011
Yeung MS, Strogatz SH (1999) Time delay in the kuramoto model of coupled oscillators. Phys Rev Lett 82(3):648
Zhang D, Snyder AZ, Shimony JS, Fox MD, Raichle ME (2010) Noninvasive functional and structural connectivity mapping of the human thalamocortical system. Cereb Cortex 20(5):1187–1194
Acknowledgements
The authors would like to thank Ana Hernando Ariza for the first image contained in this paper and the creativity, time and efforts she put into transforming the important messages of this article in graphics. The authors are particularly grateful to Prof. Jérémie Lefebvre and Prof. Micah M. Murray for their contributions, guidance and mentoring during the preparation of this article.
Funding
Open Access funding provided by Université de Lausanne. K.G. was funded by Schweizerischer Nationalfonds zur Förderung der Wissenschaftlichen Forschung Award ID: 170873. J.C. was funded by the Portuguese Foundation for Science and Technology (FCT) CEECIND/03325/2017 and by Projects UID/Multi/50026 (FCT), NORTE-01-0145-FEDER- 000013 and NORTE-01-0145-FEDER-000023. A.C. was supported by the Tiny Blue Dot Foundation. A.M. was supported by PREVIEW—Bando salute 2018—Tuscany Regional Government. A.R. was supported by NIH Grants R01NS092802, RF1AG062196 and R56AG064873. B.F. received financial support for this work by the Fondation Asile des aveugles (Grant Number 232933), a grantor advised by Carigest SA (Number 232920).
Author information
Authors and Affiliations
Contributions
K.G. and B.F. conceived the review. J.C. and A.R. contributed to section 2.2. A.M. contributed to section 2.1. A.C. entirely conceived section 4. K.G. and B.F. outlined all remaining contents and all authors contributed to the final draft and review.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Handling Editor: Jeremie Lefebvre.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
This is one of several papers published together in Brain Topography on the “Special Issue: Computational Modeling and M/EEG”.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Glomb, K., Cabral, J., Cattani, A. et al. Computational Models in Electroencephalography. Brain Topogr 35, 142–161 (2022). https://doi.org/10.1007/s10548-021-00828-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10548-021-00828-2