Abstract
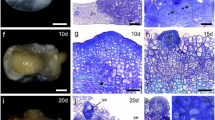
Members of the Brassicaceae family disperse their seeds through a mechanism commonly referred to as fruit dehiscence or pod shatter. Pod shatter is influenced by variations in valve margin structure and by the molecular control pathways related to valve development. Anatomical patterns of the dehiscence zone from Brassica napus L., Brassica rapa L., Brassica carinata L., and Sinapis alba L., representing fruit types differing in pod shatter resistance, were compared using histological staining. The pod shatter-susceptible plant B. napus showed an increased lignin deposition at the vascular bundle of the replum as well as an increased separation of cell layers. In pod shatter-resistant plants S. alba, B. rapa, and B. carinata, we observed two layers of lignified valve margin cells. From these four species, we isolated and identified homologs of SHATTERPROOF (SHP1, SHP2), INDEHISCENT (IND), ALCATRAZ (ALC), FRUITFULL (FUL), AGAMOUS (AG), NAC SECONDARY WALL THICKENING PROMOTING FACTOR1 (NST1), and SEEDSTICK (STK) genes involved in fruit development and pod shatter in Arabidopsis. Transcriptional analysis of these eight genes was performed by real-time polymerase chain reaction and the results demonstrate that differences in the expression patterns of the eight genes may be associated with dehiscence variation within these four species.
Similar content being viewed by others
Abbreviations
- AG :
-
AGAMOUS
- ALC :
-
ALCATRAZ
- DZ:
-
dehiscence zone
- FUL :
-
FRUITFULL
- IND :
-
INDEHISCENT
- NCBI:
-
National Center for Biotechnology Information
- NST1 :
-
NAC SECONDARY WALL THICKENING PROMOTING FACTOR1
- RT-qPCR:
-
real-time quantitative polymerase chain reaction
- SHP :
-
SHATTERPROOF
- STK :
-
SEEDSTICK
References
Arnaud, N., Girin, T., Sorefan, K., Fuentes, S., Wood, T.A., Lawrenson, T., Sablowski, R., Østergaard, L.: Gibberellins control fruit patterning in Arabidopsis thaliana. — Gene. Dev. 24: 2127–2132, 2010.
Arnaud, N., Lawrenson, T., Østergaard, L., Sablowski, R.: The same regulatory point mutation changed seed-dispersal structures in evolution and domestication. — Curr. Biol. 21: 1215–1219, 2011.
Avino, M., Kramer, E.M., Donohue, K., Hammel, A.J., Hall, J.C.: Understanding the basis of a novel fruit type in Brassicaceae, conservation and deviation in expression patterns of six genes. — Evodevo 3: 20, 2012.
Bennett, E.J., Roberts, J.A., Wagstaff, C.: The role of the pod in seed development, strategies for manipulating yield. — New Phytol. 190: 838–853, 2011.
Child, R.D., Summers, J.E., Babij, J., Farrent, J.W., Bruce, D.M.: Increaced resistance to pod shatter is associated with changes in the vaslular structure in pods of a resynthesized Brassica napus line. — J. exp. Bot. 54: 1919–1930, 2003.
Dardick, C.D., Callahan, A.M., Chiozzotto, R., Schaffer, R.J., Piagnani, M.C., Scorza, R.: Stone formation in peach fruit exhibits spatial coordination of the lignin and flavonoid pathways and similarity to Arabidopsis dehiscence. — BMC Biol. 8: 13, 2010.
Dinneny, J.R., Yanofsky, M.F.: Drawing lines and borders: how the dehiscent fruit of Arabidopsis is patterned. — Bioessays 27: 42–49, 2005.
Favaro, R.: MADS-box protein complexes control carpel and ovule development in Arabidopsis. — Plant Cell 15: 2603–2611, 2003.
Ferrándiz, C.: Regulation of fruit dehiscence in Arabidopsis. — J. exp. Bot. 53: 2031–2038, 2002.
Ferrándiz, C., Liljegren, S.J., Yanofsky, M.F.: Negative regulation of the SHATTERPROOF genes by FRUITFULL during Arabidopsis fruit development. — Science 289: 436–438, 2000.
Girin, T., Stephenson, P., Goldsack, C.M.P., Kempin, S.A., Perez, A., Pires, N., Sparrow, P.A., Wood, T.A., Yanofsky, M.F., Østergaard, L.: Brassicaceae INDEHISCENT genes specify valve margin cell fate and repress replum formation. — Plant J. 63: 329–338, 2010.
Gonzalez-Reig, S., Ripoll, J.J., Vera, A., Yanofsky, M.F., Martínez-Laborda, A.: Antagonistic gene activities determine the formation of pattern elements along the mediolateral axis of the Arabidopsis fruit. — PLoS Genet. 8: e1003020, 2012.
Hua, S., Shamsi, I.H., Guo, Y., Pak, H., Chen, M., Shi, C., Meng, H., Jiang, L.: Sequence, expression divergence, and complementation of homologous ALCATRAZ loci in Brassica napus. — Planta 230: 459–503, 2009.
Kadkol, G.P.: Brassica shatter-resistance research update. - In: Proceedings of the 16th Australian Research Assembly on Brassicas. Pp. 104–109. Ballarat, Victoria 2009.
Lenser, T., Theißen, G.: Conservation of fruit dehiscence pathways between Lepidium campestre and Arabidopsis thaliana sheds light on the regulation of INDEHISCENT. — Plant J. 76: 545–556, 2013.
Lewis, M.W., Leslie, M.E., Liljegren, S.J.: Plant separation: 50 ways to leave your mother. — Curr. Opin. Plant Biol. 9: 59–65, 2006.
Liljegren, S.J., Ditta, G.S., Eshed, Y., Savidge, B., Bowman, J.L., Yanofsky, M.F.: SHATTERPROOF MADS box genes control seed dispersal in Arabidopsis. — Nature 404: 766–770, 2000.
Liljegren, S.J., Roeder, A.H., Kempin, S.A., Gremski, K., Østergaard, L., Guimil, S., Reyes, D.K., Yanofski, M.F.: Control of fruit patterning in Arabidopsis by INDEHISCENT. — Cell 116: 843–853, 2004.
Livak, K.J., Schmittgen, T.D.: Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. — Methods 25: 402–408, 2001.
Martinez-Andujar, C., Martin, R.C., Nonogaki, H.: Seed traits and genes important for translational biology ? highlights from recent discoveries. — Plant Cell Physiol. 53: 5–15, 2012.
Mitsuda, N., Ohme-Takagi, M.: NAC transcription factors NST1 and NST3 regulate pod shattering in a partially redundant manner by promoting secondary wall formation after the establishment of tissue identity. — Plant J. 56: 768–778, 2008.
Mühlhausen, A., Lenser, T., Mummenhoff, K., Theißen, G.: Evidence that an evolutionary transition from dehiscent to indehiscent fruits in Lepidium (Brassicaceae) was caused by a change in the control of valve margin identity genes. — Plant J. 73: 824–835, 2013.
Mummenhoff, K., Polster, A., Mühlhausen, A., Theißen, G.: Lepidium as a model system for studying the evolution of fruit development in Brassicaceae. — J. exp. Bot. 60: 1503–1513, 2009.
Ogawa, M., Kay, P., Wilson, S., Swain, S.M.: ARABIDOPSIS DEHISCENCE ZONE POLYGALACTURONASE1 (ADPG1), ADPG2, and QUARTET2 are polygalacturonases required for cell separation during reproductive development in Arabidopsis. — Plant Cell 21: 216–233, 2009.
Østergaard, L., Kempin, S.A., Bies, D., Klee, H.J., Yanofsky, M.F.: Pod shatter-resistant Brassica fruit produced by ectopic expression of the FRUITFULL gene. — Plant Biotechnol. J. 4: 45–51, 2006.
Østergaard, L., King, G.J.: Standardized gene nomenclature for the Brassica genus. — Plant Methods 4: 10, 2008.
Pinyopich, A., Ditta, G.S., Baumann, E., Wisman, E., Yanofsky, M.F.: Unraveling the redundant roles of MADS-box genes during carpel and fruit development. — Nature 424: 85–88, 2003.
Rajani, S., Sundaresan, V.: The Arabidopsis myc/bHLH gene ALCATRAZ enables cell separation in fruit dehiscence. — Curr. Biol. 11: 1914–1922, 2001.
Roeder, A.H., Yanofsky, M.F.: Fruit development in Arabidopsis. - In: Somerville, C., Meyerowitz, E. (ed.): The Arabidopsis book. Pp 1–50. American Society of Plant Biologists, Rockville 2006.
Seymour, G.B., Østergaard, L., Chapman, N.H., Knapp, S., Martin, C.: Fruit development and ripening. — Annu. Rev. Plant Biol. 64: 219–241, 2013.
Sorefan, K., Girin, T., Liljegren, S.J., Ljung, K., Robles, P., Galván-Ampudia, C.S., Offringa, R., Friml, J., Yanofsky, M.F., Østergaard, L.: A regulated auxin minimum is required for seed dispersal in Arabidopsis. — Nature 459: 583–586, 2009.
Spence, J., Vercher, Y., Gates, P., Harris, N.: Pod shatter in Arabidopsis thaliana, Brassica napus and B. juncea. — J. Microscopy 181: 195–203, 1996.
Summers, J.E., Bruce, D.M., Vancanneyt, G., Redig, P., Werner, C.P., Morgan, C., Child, R.D.: Pod shatter resistance in the resynthesized Brassica napus line DK142. — J. agr. Sci. 140: 43–52, 2003.
Wang, R., Ripley, V.L., Rakow, G.: Pod shatter resistance evaluation in cultivars and breeding lines of Brassica napus, B. juncea and Sinapis alba. — Plant Breed. 126: 588–595, 2007.
Wu, H., Mori, A., Jiang, X., Wang, Y., Yang, M.: The INDEHISCENT protein regulates unequal cell divistions in Arabidopsis fruit. — Planta 224: 971–979, 2006.
Zhao, Q., Dixon, R.A.: Transcriptional networks for lignin biosynthesis, more complex than we thought? — Trends Plant Sci. 16: 227–233, 2011.
Author information
Authors and Affiliations
Corresponding author
Additional information
Acknowledgments: This work was supported by the National Natural Science Foundation of China (31370258).
Electronic supplementary material
Rights and permissions
About this article
Cite this article
Zhang, Y., Shen, Y.Y., Wu, X.M. et al. The basis of pod dehiscence: anatomical traits of the dehiscence zone and expression of eight pod shatter-related genes in four species of Brassicaceae . Biol Plant 60, 343–354 (2016). https://doi.org/10.1007/s10535-016-0599-1
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10535-016-0599-1