Abstract
Sturgeon populations are declining worldwide and are the target of extensive conservation efforts. Addressed in several pieces of legislation, sturgeons have received considerable attention as flagship or umbrella species. Despite the need for a better understanding of the distribution and population status, the use of traditional sampling methods failed in the past, thereby hampering reliable assessments, a prerequisite for conservation. Here, we describe the development and application of an environmental DNA (eDNA) metabarcoding approach for detecting rare sturgeons in large rivers. Exemplarily, we developed a reference database for five native Danube sturgeons (Acipenser stellatus, Acipenser gueldenstaedtii, Acipenser ruthenus, Acipenser nudiventris, and Huso huso) and two non-native species (Acipenser baerii and Acipenser transmontanus), assessed these ex situ, and used eDNA as a detection tool along the entire length of the Danube (Europe, ~ 2850 km) and major tributaries. In ex situ analyses, all assays yielded positive amplifications for the assessed sturgeon species. In the Danube, the presence of A. ruthenus was confirmed at 14 of 29 sites (48.3%), and in 2 of 18 tributary sites (11.1%), providing the first comprehensive large-scale biogeographical snapshot of this species. Relative number of reads assigned to A. ruthenus varied between 0 and 2.5%, with sites registering positive detections being clustered in 3 sections of the Danube. Our findings enabled us to confirm the advantages of eDNA monitoring over traditional sampling methods for comprehensive whole-river snapshot studies of sturgeons conducted on a large geographical scale, and therefore we consider it to be a promising approach for application in conservation measures, fisheries management, scientific studies, and adaptive management plans for sturgeons on a global scale.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Sturgeons are amongst the most ancient of extant fishes (traced back to approximately 200 MYA) and considered one of the most endangered group of vertebrates worldwide, with more than 85% of the species being classified as either endangered or threatened (IUCN 2021). The precipitous decline in populations globally is closely linked to overharvest and the loss of habitats associated with alterations in river functionality (e.g. Luk'Yanenko et al. 1999). Facilitating recovery of the dwindling sturgeon populations requires an in-depth understanding of their ecology and the main factors causing decline. Currently, however, there is still relatively little global scientific information available regarding the autecology and population status of many sturgeon species, which can be ascribed to both the lack of suitable sampling methods for their detection and the small sizes of remnant populations (Jarić et al. 2018). Given the longevity of these species, functional extinction may occur decades before they actually become extinct, and thus reliable methods that can be used to confirm presence or absence, but also to assess species abundance and population structure, are required to enable the timely initiation of rescue programs. Furthermore, conservation efforts are largely dependent on the provision of cost-effective precise data within an ecologically and politically actionable timeframe (Thomsen and Willerslev 2015).
Traditional sampling methods often require concerted efforts and subsequently high expenditure in order to gain sufficiently valuable data on species that may be on the brink of extinction. This is particularly applicable in the case of large rivers, for which there are few standardized sampling methods, owing to the often extremely variable environmental conditions and the fact that all sampling techniques have associated limitations and biases (Casselman et al. 1990; Hill et al. 2005). For example, typically only those sections of rivers that have low flow are suitable for the use of bottom-set stationary gear, such as gill nets, trap nets, long lines, and drifting gillnets, and consequently only these sections tend to be sampled. Furthermore, boat electrofishing is limited by water transparency and the depth limitation of the electric field, thereby restricting its application to shallow littoral shorelines (CEN 2003). Given the various difficulties associated with sampling, the probability of detecting these rare species using traditional methods is low and challenging (MacKenzie et al. 2005; Szalóky et al. 2014). Moreover, all the aforementioned methods are invasive or harmful to some extent, which is a particularly important consideration in the case of rare and protected species on the brink of extinction. Additionally, for assessments of the population status of riverine species, time-limited systematic surveys are required, and quantitative monitoring of fish assemblages remains a difficult and costly task, particularly in large rivers (Zajicek and Wolter 2018). Consequently, novel and innovative techniques are required for sampling rare species in large rivers.
An alternative approach for monitoring rare or elusive species in such environments is the use of environmental DNA (eDNA). Using this approach, water samples, as opposed to physical specimens, can be used to identify those species that have recently been present in a local environment, with genetic material consisting of intracellular and extracellular DNA (e.g., skin cells, intestinal cells, scales, and/or mucus) being obtained directly from environmental samples (Taberlet et al. 2012). Recently, this approach has frequently been used to detect rare and imperiled freshwater fishes and entire fish communities (e.g., Czeglédi et al. 2021; Hänfling et al. 2016; Pont et al. 2018)). Importantly, sampling free eDNA in water is non-invasive and facilitates species detection without the need to capture individual specimens, thereby avoiding handling-related stress and mortality, which is of particular importance when monitoring species that are either rare or elusive. Moreover, the technique is potentially faster, less expensive, and less destructive than traditional sampling methods (Thomsen et al. 2012). In addition, a significant correlation has been observed between species relative abundance and the number of standardized reads, enabling a quantitative estimate of fish assemblage structures (hereafter referred to as relative species abundance) (Pont et al. 2018; Rourke et al. 2022).On the other hand, there are some limitations in the applicability and informative power of the method, such as false negatives and false positives, lack of information on population structure or the exact location of a sturgeon (see also Discussion). To date, sturgeons have been targeted only in a very limited number of eDNA studies in the USA (Mobile River Basin (Pfleger et al. 2016), Sacramento River (Bergman et al. 2016), and Hudson River (Stoeckle et al. 2017)), Canada [Winnipeg River (Yusishen et al. 2020)], and China [Yangtze River (Xu et al. 2018)]. In contrast, there have been no eDNA-based studies conducted in the River Danube for any of its native species. Moreover, given the aforementioned limitations, the use of traditional sampling methods has resulted only in very limited catches of Danube sturgeons and published data are sparse (e.g. Bartosiewicz et al. 2008; Paraschiv et al. 2006; Vassilev and Pehlivanov 2003).
The Danube River Basin is home to 6 native sturgeon species. Historically, these have ranged with decreasing abundance from the Black Sea to the subalpine region, also entering major tributaries (Holčík 1989). The beluga sturgeon (Huso huso) and the stellate sturgeon (Acipenser stellatus) still reproduce on a very small scale in the Lower Danube, although since 2018, increasing numbers of natural hybrids with the more abundant sterlet (Acipenser ruthenus) have been detected, tending to indicate further reductions in spawning animals (Ionescu, pers. comm.). Among the other species, available data for the Russian sturgeon (Acipenser gueldenstaedtii) indicate a cessation of active reproduction in 2010, and thus a high risk of future extinction (Friedrich et al. 2018; Lenhardt et al. 2006), whereas the sterlet is encountered in the Middle and Lower Danube. In the Upper Danube only a single reproductive population is known near the Austrian–German border, with occasional catches in other sections (Friedrich et al. 2019). The ship sturgeon (Acipenser nudiventris) is considered to be functionally extinct in the Danube (Jarić et al. 2016; Reinartz and Slavcheva 2016) as only 3 specimens were caught in the period from 2000 to 2010 in the Middle Danube. It is, nevertheless, currently unclear as to whether isolated single old individuals remain. The European sturgeon (Acipenser sturio) is considered extinct in the Danube and the Black Sea Basin (Bacalbasa-Dobrovici and Holcik 2000).
Currently, species of Danube sturgeon are being targeted for protection by several legislative documents and conservation directives, and with the exception of the sterlet (vulnerable), all species have been listed as critically endangered on the International Union for Conservation of Nature (IUCN) Red List of threatened species. Furthermore, these species are listed in different annexes of the Cites Convention, Habitat Directive, and Bern and Bonn Conventions. The action plans and projects of several organizations, including, the EU Strategy for the Danube Region (EUSDR), International Commission for the Protection of the Danube River (ICPDR), Danube Sturgeon Task Force (DSTF), World Wide Fund For Nature (WWF), the Pan-European Action Plan for Sturgeons (PANEUAP), and various EU-funded LIFE and INTERREG projects, have endorsed sturgeons as flagship species, as these fish are notably sensitive to environmental pressures. Accordingly, they can function as valuable indicators of healthy rivers, and also serve as umbrella species for fostering freshwater conservation (Carrizo et al. 2017).
To facilitate sustainable conservation initiatives, it is of the utmost importance to gain a reliable understanding of the present status and development of remnant populations and sound study results providing evidence-based data for making water management decisions (Meulenbroek et al. 2020). In the case of the Danube, however, suitable standardized methods for assessing the status of sturgeon populations have yet to be agreed upon and put in place. Indeed, monitoring the populations of these fish presents innumerable challenges in long, large international rivers such as the Danube, which has a total length of > 2800 km, passes through or borders 10 countries, and discharges a volume of more than 6400 m3 s−1 (Sommerwerk et al. 2009).
In this study, we aimed to (1) develop a reference database for several species of Danube sturgeons, (2) validate eDNA metabarcoding primers ex-situ, and (3) utilize eDNA as a new detection tool along the entire length of the River Danube (~ 2800 km) and its major tributaries, in order to provide information on the distribution and relative abundance of species in the community.
Methods
Study site and in situ eDNA sampling
For the purposes of the present study, we selected a total of 47 sites covering the entire Danube catchment. The 29 sampling sites along the Danube itself were selected to provide a reasonably comparable distance between sites (average: 99.2 km, Sx: 26.0 km, range: 38–149 km). By adopting sampling site distances of this scale, we could effectively minimize the potential influence of eDNA transported from the site immediately upstream (Pont et al. 2018). Furthermore, we ensured that no sampling sites were located closely downstream of the confluence of a major tributary. During the same period, we collected samples from 18 tributaries at distances of between 1 and 55 km upstream from their confluence with the Danube. Sites were sampled between June 29 and July 19, 2019, with the exception being a single site near Vienna (sampled August 6, 2019). Owing to no or low DNA amplification from samples at the Inn River site, this site was re-sampled in May 2020, and for site “Hainburg”, samples collected in July 2017 were used. Further information regarding the sampling sites (sampling date, location along the Danube, coordinates, discharge and physico-chemical water paramters) is presented in Table S1. Sampling was conducted under conditions of approximately mean discharge. At each site, 2 surface samples (less than 50 cm depth) were collected using a peristaltic pump, either by wading or from a boat moving from shore to shore to facilitate an integrated sampling of the river cross-section. The water was filtered (VigiDNA 0.45-μm crossflow filtration capsule; SPYGEN), using sterile disposable tubing. Having completed filtration, the water in the capsule was drained and the capsule was filled with 80 mL of preservation buffer CL1 (SPYGEN) to prevent eDNA degradation. The mean filtration time per sample and the mean volume of water filtered were 22.34 min (3 to 40 min) and 28.73 L (3 to 40 L), respectively, depending on the clogging rate of the filtration capsule.
eDNA extraction, PCR amplification, and analysis
The procedure used for the eDNA metabarcoding workflow (extraction, amplification using “teleo” primers, high-throughput sequencing, and bioinformatic analysis) followed the protocol described by Pont et al. (2018). “Teleo” primers (Sequence 5′–3′: forward: ACACCGCCCGTCACTCT; reverse: CTTCCGGTACACTTACCATG) amplify a short fragment (roughly 60 bp) at the end of the 12S rRNA region (Valentini et al. 2016). The extracted eDNA was PCR amplified, with 12 replicate reactions being performed for each sample. A corresponding 12 libraries were prepared using the Fasteris MetaFast protocol and 12 paired-end sequencings (2 × 125 bp) were carried out in a Miseq sequencer (Illumina) at Fasteris facilities, using a Miseq Kit v3 (Illumina) following the manufacturer's instructions. To monitor for potential contaminants, 11 negative extraction controls and seven negative PCR controls (ultrapure water) were amplified with 12 replicates and sequenced in parallel with the aforementioned samples. Sequence reads were analyzed using programs implemented in the OBITools package (Boyer et al. 2016). The forward and reverse reads were assembled using the ILLUMINAPAIREDEND program, based on a minimum score of 40 and retrieving only joined sequences. Subsequently, we assigned the reads to each sample using NGSFILTER software and a separate data set was created for each sample by splitting the original data set into several files using OBISPLIT. Thereafter, we analyzed each sample individually prior to merging the taxon list for the final ecological analysis. Strictly identical sequences were clustered together using OBIUNIQ. Sequences shorter than 20 bp, or with fewer than 10 (or labeled “internal” by the OBICLEAN program) occurrences were excluded. The taxonomic assignment of molecular operational taxonomical units was performed using the ECOTAG program, based on a local “Sturgeon” reference database (see later), the “Danubian” database, obtained from (Pont et al. 2022), the database obtained from Valentini et al. (2016), and the sequences extracted from the release 142 (standard sequences) of the ENA database (http://www.ebi.ac.uk/ena). Given the incorrect assignment of a few sequences to the sample due to tag-jumps (Schnell et al. 2015), all sequences with a frequency of occurrence < 0.001 per sequence and per library were discarded. The data thus obtained were curated for Index-Hopping (MacConaill et al. 2018) with a threshold empirically determined for each sequencing batch using experimental blanks (i.e., combinations of tags not present in the libraries) for a given sequencing batch between libraries. Relative species abundance refers to relative number of standardized reads of all fish species as described in Pont et al. (2018). Total number of reads, % positive PCR and number of reads for Acipenser ruthenus are given in Table S2. A total of 60 taxa known to occur in the Danube River Basin were detected. 48 taxa were assigned at the species level, while 12 taxa assigned at a higher taxonomic level corresponded to a potential of 26 known Danubian species, resulting in a maximum number of 74 species detected. A full list of all species per site is available in Fig. 1 in Pont et al. (2021); and in Suppl. Figure 1 in Pont et al. (2022). This value was comparable to the total number of 71 species captured in the TEF survey conducted during the same period (Bammer et al. 2021).
Reference database construction and ex situ validation
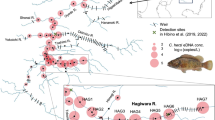
Detections of Acipenser ruthenus based on eDNA water samples collected from the Danube and selected tributaries. Green boxes indicate clusters of positive sites in 3 sections of the Danube (also see Fig. 2)
All reference specimens (Acipenser stellatus, Acipenser gueldenstaedtii, Acipenser ruthenus, Acipenser nudiventris, Huso huso, Acipenser baerii, and Acipenser transmontanus) were provided by the Institute of Hydrobiology and Aquatic Ecosystem Management, University of Natural Resources and Life Sciences, Vienna, either from wild catches or from a hatchery. Total DNA was extracted from 10 mg of muscle tissue, following the protocol described in Valentini et al. (2016). The extracted DNA was amplified using the eDNA metabarcoding protocol with “teleo” primers and was sequenced using a Miseq sequencer at Fasteris facilities. The sequences thus obtained were analyzed using the ObiTools package following the same protocol used for eDNA samples, excluding the taxonomical assignation step. The most abundant sequence was retrieved for reference database construction.
At a hatchery station on the Danube Island in Vienna (https://life-sterlet.boku.ac.at/index.php/the-project.html), 3 A. baerii (total biomass 180 g), 3 A. gueldenstaedtii (180 g), 2 A. nudiventris (210 g), 30 A. ruthenus (7500 g), and 2 H. huso (140 g) specimens have been housed in a circular tank (volume = 0.9 m3, water flow through 0.5 L/s) as a part of a conservation breeding program. Using water samples collected from the tank, we evaluated our sampling protocols and the reliability of the developed assay. A single 28-L water sample was collected as described above with a 20-min filtration time. eDNA metabarcoding analysis was performed using the aforementioned procedure.
Results
All sequences obtained for reference database construction proved to be effective in distinguishing the respective target sturgeon species (Table 1). The identity between species was found to vary from 98% between A. stellatus and A. ruthenus (a 1 base difference) to 92% between A. stellatus and A. transmontanus (a 5 base difference). One species, A. gueldenstaedtii, was found to have 2 haplotypes differing by a single base.
In the ex situ experiment, all replicates showed a positive amplification, regardless of species and fish abundance and biomass (Table 2). For species with a low number of individuals (2 or 3) and biomass (140–210 g), we obtained between 2060 and 3780 DNA reads, whereas for the more numerous A. ruthenus (30 ind., 7500 g), we obtained more than 150,000 DNA reads. Interestingly, we also detected the presence of A. transmontanus, a single specimen of which had previously been present in the tank, although at the time of sampling, had been removed more than a week previously. However, only 2 positive replicates and 25 reads were obtained.
With respect to the in-situ monitoring, we confirmed positive amplifications for A. ruthenus from 14 of the 29 sampling sites (48.3%) along the Danube, whereas samples from 2 of the 18 tributary sampling sites (11.1%), the rivers Inn and Tisza (Fig. 1), yielded positive identifications. Figure 2 presents data obtained for the relative species abundance and percentage of positive PCR detections for A. ruthenus along the River Danube from sea to source. Notably, sites registering a positive identification were found to cluster in 3 discrete sections of the Danube:
-
(1)
The Danube delta (mainly Kilia Arm at Vylkove and Reni to Giurgeni);
-
(2)
Upstream of the Iron Gate I (mainly at Tekija and Banatska Palanka); and
-
(3)
From Budapest to Gabčíkovo.
Where present, A. ruthenus accounted for an average relative species abundance of 0.45%, and 0.16% when including all sites. At only a single site did the relative species abundance exceed 1%, namely, in the Danube Delta region at river kilometer (rkm) 18 (2.5%). The specific numbers of positive PCR replicates, as well as the total numbers of DNA sequences, are presented in Table S2.
The presence of A. stellatus was detected in samples collected from the most downstream of the sampling sites (Kilia Arm Vylkove), whereas evidence of A. gueldenstaedtii was obtained only at the River Inn site. However, no positive amplification of the other targeted sturgeon species were detected in the field survey.
Discussion
The primers pairs used in this study are able to distinguish all tested Acipenseridae species. In addition, analysis of samples collected ex situ, based on teleo markers and an eDNA metabarcoding approach, are sufficiently sensitive for detecting the occurrence of the endangered Danube sturgeons. The integrative sampling strategy we adopted in situ confirmed that this approach is suitable for the detection of sterlet and a first comprehensive large-scale biogeographic snapshot is presented.
Reports regarding the detection of sturgeon species are generally very limited in terms of temporal and spatial coverage. This is perhaps not surprising, given that sturgeons are typically bottom-dweller that are nowadays exceptionally rare species in many large river systems, and are exceedingly difficult to detect using traditional monitoring methods. Consequently, most of the currently available data are derived from small-scale scientific studies (sometimes conducted over distances of only several hundred meters). Although fisheries data can provide a broader picture on larger scales, they are of limited value in certain respects (e.g. missing information on catch per unit effort, seasonal changes, gear consideration, illegal fisheries) (Guy and Brown 2007). To the best of our knowledge, the only systematic basin-wide traditional fish assessments of the Danube Basin have been the Joint Danube surveys (conducted in summer 2007, 2013, and 2019) (Bammer et al. 2021, 2015; Wiesner et al. 2008). However, despite the high sampling effort of more than 2500 electro-fished stretches covering close to 3 Mio m2, only 3 sturgeon individuals at 2 sites have been caught. The use of electrified benthic frame trawls and trammel nets did, nevertheless, enable the capture of sturgeons from 3 additional sampling sites. These findings accordingly serve to highlight that traditional sampling methods have hitherto failed to provide a comprehensive picture regarding the distribution of sturgeon species across a large geographical scale along a longitudinal profile of the Danube. eDNA-based surveys have the potential to overcome these limitations in aquatic systems that are generally unamenable to monitoring using traditional methods (Jerde 2021), and can serve as an initial first step in identifying key habitats and sampling sites for the targeted monitoring of population sizes and recruitment.
However, although there is a growing body of literature highlighting the various merits of eDNA sampling compared with traditional sampling (e.g. Czeglédi et al. 2021; Fediajevaite et al. 2021; Hänfling et al. 2016; Pont et al. 2018; Valentini et al. 2016), particularly with respect to large water bodies and rare species, the technique does, nevertheless, have certain drawbacks and restrictions. Capture-based methods have a distinct advantage in that they can provide information relating to population structure and the condition of individual fish, as well as the certainty of having physical specimens (Jerde 2021). In particular, information on population structure is of great importance for early detection of the risk of functional extinction. Interpretation of the data must also take into account that mtDNA markers, such as those used in the present study, cannot identify hybrids. A further limitation is that identification of the presence of a species does not necessarily pinpoint the exact location of fish, particularly in the case of flowing water bodies. Under conditions of hydrological transport, eDNA essentially functions as a carrier of material containing genetic information on the biodiversity of upstream catchments (Deiner et al. 2016). Moreover, detection distances can vary significantly from a few kilometers in small streams to more than 100 km in large rivers (Pont et al. 2018 and references therein). In this regard, this limitation could also be viewed as an advantage, as longer river sections can be integrated in a single sample. A further issue of particular importance in the context of surveys conducted for rare, low-density, or possibly extinct animals is the occurrence of false positive (see further Darling et al. 2021) and false negative readings. For example, false positives can arise in instances in which although target DNA is present in a sample, no living organisms are present in the sampled system. In this case, eDNA could plausibly be derived via contamination from external sources, such as sewage and wastewater discharge or effluents from aquaculture farms upstream of the sampling site (Rees et al. 2014). This is potentially applicable in the case of sturgeon species, particularly in the Lower Danube catchment, in which sturgeon are traded in fish markets, served in restaurants, and cultured in fish farms, and hence it is conceivable that DNA originating from these sources might enter river systems. Conversely, false negatives (e.g., organisms in the system but no detectable DNA) could, for example, arise if the amount of target DNA falls below the limit of detection, DNA of non-target species interfere with the reaction, inhibitors are present in the sample, or in the case of insufficient sampling effort (Rees et al. 2014). The effect of eDNA concentration dilution is an issue of particular concern with respect to the detection of rare species in large rivers.
However, when using eDNA metabarcoding in such environments, there are several strategies that can be adopted to potentially enhance the probability of detection, notably, increasing sample replication at either one or both of the following 2 levels: sample collection and molecular replicates (e.g., number of PCR assays in the laboratory) (Ficetola et al. 2015). In the case of detecting rare eDNA in samples with very poor molecular detection probability, an increase in molecular replicates is particularly advisable (Erickson et al. 2019). Accordingly, in this study we performed 2 replicate samplings and 12 PCR replicates (for each replicate sample), which is considered a more than sufficient replication effort for species with low detection probability (Ficetola et al. 2015). Increasing the number of reference specimens could potentially increase the number of haplotypes per species and therefore increase the probability of detection. Collecting a larger number of samples would also be beneficial and is particularly important in the context of metabarcoding studies that set minimum acceptance thresholds for the number of reads considered indicative of a true positive signal. To overcome this issue, we performed an integrative sampling strategy in space (an entire section of the river) and time (approx. half an hour) and by collecting relatively large volumes of water (28.73 L per sample on average). Systematically collecting from a large number of sampling sites in this way would certainly contribute to maximizing the likelihood of detection (Cantera et al. 2019). Furthermore, when practical, we would recommend sampling at sites with low stream flows and at different times throughout the year.
Among the sturgeon species known to inhabit the Danube, the sterlet (A. ruthenus) is the only species that is encountered occasionally throughout the entire length of the river (Friedrich et al. 2019). However, despite being of a certain economic value in the Middle and Lower Danube catchments (Guti 2008; Vassilev and Pehlivanov 2003), the current status of the population remains almost completely unknown, although the consensus among most authors is that stocks are declining (e.g. Bacalbasa-Dobrovici 1991; Paraschiv et al. 2006) and aging (Kubala et al. 2021). In the present study, approximately half of the samples collected from the Danube yielded a positive signal for the sterlet, and notably, we identified 3 areas with higher densities of this species, as indicated by a high frequency of detection and a high number of reads (namely, the Danube Delta, Iron Gate to Belgrade, and Budapest to Gabčíkovo sections of the river). The highest rates of sterlet detection were obtained for the Delta region of the Lower Danube (rkm 0–862) with a subsequent reduction in detection until rkm 235. An absence of positive signals further upstream between rkm 375 and 700 would tend to be indicative of a discontinuous longitudinal distribution of populations within River Danube. However, genetic analysis has indicated the occurrence of gene flow and a low level of sub-structuring within the Danube (Cvijanović et al. 2017; Reinartz et al. 2011). The aforementioned stretch of the Danube has, nonetheless, experienced a marked reduction in sterlet stocks over the past few decades (Vassilev and Pehlivanov 2003), and thus further research is needed to explain these results. In contrast, with the exception of a single site at rkm 1560, sterlet were successfully detected at all sampling points in the Middle Danube (rkm 943–1790), with the highest relative species abundances being recorded from the stretches between Iron Gates and Belgrade and from Budapest to Gabčíkovo. Even so, recent massive declines have been reported for this section of the Danube, which have mainly been attributed to the destruction of important spawning habitats during construction of the Gabčikovo Dam (Guti 2008). In the Upper Danube (rkm > 1790) sterlets were detected at only 2 sampling sites, namely, downstream of Vienna (rkm 1920) in the tailwater of the Freudenau hydropower plant and in Deggendorf (rkm 2282). Since the twentieth century, it appear that sterlets have been present in the Upper Danube in only low numbers within a few fragmented populations (Friedrich et al. 2019), most of which are actively supplemented by periodic re-stocking (Friedrich 2018). A similar practice is in place at Vienna, where a remnant wild population of an estimated few dozen individuals is actively maintained by re-stocking (Friedrich et al. 2016). The sample at Jochenstein with a known small reproductive population (Zauner 1997) did not detect the species in this study. Although there are historical reports of the presence of sterlet at the most upstream site at Deggendorf (Reinartz 2008 references therein; e.g. Streibl 1920), their occasional detection by commercial fisheries in this area can probably be ascribed to stocking activities. It should also be noted that the results consider only one point in time and may be influenced by seasonal effects such as seasonal migration patterns and reproduction.
With respect to the other sturgeon species, we successfully detected the targeted species in water samples collected in an ex situ environment (see Table 2). Notably, in these analyses, we also detected the presence of A. transmontanus, even though this species had been removed from the sampled tank more than a week prior to collecting water samples. In this regard, it has been established that eDNA can persist and remain detectable by PCR for between a day and approximately one month after the removal of animals, depending on environmental conditions (Barnes et al. 2014; Dejean et al. 2011). However, our analysis of field samples revealed only single occurrences of A. stellatus (Danube Delta) and A. gueldenstaedtii (River Inn), the latter of which we suspect originated from rearing ponds upstream of the sampling site. The fact that none of the other target species were detected, would thus tend to indicate that these are either practically absent from the sampling sites or that the amounts of DNA in samples were below the limit of detection of our current methodology, both of which are plausible. Acipenser nudiventris is considered to be functionally extinct in the Danube (Reinartz and Slavcheva 2016; Simonovic et al. 2005) and all other anadromous species (H. huso, A. stellatus, and A. gueldenstaedtii) are currently listed as critically endangered, occurring in the Lower Danube in only small numbers up to the Iron Gate site (Friedrich et al. 2018). A further factor that might account for our inability to detect these rare anadromous species is the timing of sampling. For Ponto‐Caspian sturgeons, there are 4 known patterns of migration with differing peaks that occur either in spring or during fall (Berg 1934; Holčík 1989). However, whereas sampling in July (summer, as conducted in the present study) may not be conducive to detecting the adults of early-spawning species such as the beluga sturgeon, this timing should be ideal for detecting the presence of either the young‐of‐the‐year and/or mature adults of all the targeted species.
One consistent request by several organizations, action plans, and projects that have targeted sturgeon conservation is the establishment of a permanent and standardized monitoring program (e.g. Friedrich 2018; Friedrich et al. 2018; ICPDR 2018; Sandu et al. 2013), which would contribute to documenting changes in population dynamics for adaptive management. Such a permanent monitoring network could be readily implemented if based on repeated eDNA sampling campaigns. In addition, this would also facilitate further studies on seasonal migration patterns from the marine environment into rivers (see Stoeckle et al. 2017), as well as assessments of the efficacy of population support actions and the characterization of key habitats, such as spawning, overwintering, or nursery areas, that could subsequently be investigated and, if necessary, protected. Devising effective sampling methods for monitoring contributes to meeting one of the urgent priority needs of supporting the informed management of freshwater biodiversity, which is a vital step in enhancing coordinated action for its sustainable management and conservation (Maasri et al. 2021).
In conclusion, in this study, we demonstrate the practical utility of the eDNA metabarcoding approach as a tool for monitoring sturgeon species in large rivers, as illustrated by our survey of the entire Danube. Moreover, we present the first comprehensive whole-river snapshot study of Acipenser ruthenus conducted on a large geographical scale. Given certain limitations of our current methodology, the sampling strategy will need to be modified for the assessment of other endangered sturgeon species to counteract the dilution effect of very low eDNA concentrations. An in-depth understanding of species distribution and population dynamics is essential for developing adaptive conservation management plans, and in this regard, the benefits of an eDNA approach for conservation efforts, fisheries management, and scientific studies are numerous, particularly for rare bottom-dwelling species inhabiting large rivers. Capture-based methods have the distinct advantage of being able to provide information on population structure, which will make them indispensable also in the future. However, techniques based on short species-specific eDNA fragments are potentially more sensitive than traditional survey methods, as well as being more cost-effective, non-invasive, and facilitating time-limited coverage of large geographical areas, thereby enabling the implementation of conservation measures within an ecologically and politically actionable time scale.
Data availability
The datasets generated during and/or analysed during the current study are available from the corresponding author on reasonable request.
Change history
25 November 2022
A Correction to this paper has been published: https://doi.org/10.1007/s10531-022-02491-w
References
Bacalbasa-Dobrovici N (1991) Statut des differentes especes esturgeons dans le Danube Roumain: problemes lies a leur maintenance Acipenser. Cemagref Publ, Antony, pp 185–192
Bacalbasa-Dobrovici N, Holcik J (2000) Distribution of Acipenser sturio L., 1758 in the Black Sea and its watershed. BOLETIN-INSTITUTO ESPANOL DE OCEANOGRAFIA 16:37–42
Bammer V, György A, Pehlivanov L, Schabuss M, Szaloky Z, Zornig H (2015) Joint Danube Survey 3 - A Comprehensive Analysis of Danube Water Quality - Chapter 9 FISH. ICPDR International Commission for the Protection of the Danube River, Vienna
Bammer V, Apostolou A, Bulat D, Dumitrascu OC, Effenberger M, Erős T, Hortic S, Simonović P (2021) Joint Danube Survey 4 - Scientific Report: a Shared Analysis of the Danube River - Chapter 5 FISH. ICPDR International Commission for the Protection of the Danube River, Vienna
Barnes MA, Turner CR, Jerde CL, Renshaw MA, Chadderton WL, Lodge DM (2014) Environmental conditions influence eDNA persistence in aquatic systems. Environ Sci Technol 48:1819–1827
Bartosiewicz L, Bonsall C, Şişu V (2008) Sturgeon fishing in the middle and lower Danube region. The Iron Gates in Prehistory. Archaeopress, Oxford, pp 39–54
Berg L (1934) Vernal and hiemal races among anadromous fishes. Izvestya Akademii Nauk SSSR 5:711–732
Bergman PS, Schumer G, Blankenship S, Campbell E (2016) Detection of adult green sturgeon using environmental DNA analysis. PLoS ONE 11:e0153500
Boyer F, Mercier C, Bonin A, Le Bras Y, Taberlet P, Coissac E (2016) obitools: A unix-inspired software package for DNA metabarcoding. Mol Ecol Resour 16:176–182
Cantera I, Cilleros K, Valentini A, Cerdan A, Dejean T, Iribar A, Taberlet P, Vigouroux R, Brosse S (2019) Optimizing environmental DNA sampling effort for fish inventories in tropical streams and rivers. Sci Rep 9:1–11
Carrizo SF, Jähnig SC, Bremerich V, Freyhof J, Harrison I, He F, Langhans SD, Tockner K, Zarfl C, Darwall W (2017) Freshwater megafauna: flagships for freshwater biodiversity under threat. Bioscience 67:919–927
Casselman JM, Penczak T, Carl L, Mann RH, Holcik J, Woitowich WA (1990) An evaluation of fish sampling methodologies for large river systems. Pol Arch Hydrobiol 37:521–551
CEN (2003) 14011: 2003 Water analysis—fishing with electricity for wadable and non-wadable rivers. European Committee for Standardization
Cvijanović G, Adnađević T, Jarić I, Lenhardt M, Marić S (2017) Genetic analysis of sterlet (Acipenser ruthenus L.) populations in the Middle and Lower Danube sections. North-West J Zool 13(1):34–43
Czeglédi I, Sály P, Specziár A, Preiszner B, Szalóky Z, Maroda Á, Pont D, Meulenbroek P, Valentini A, Erős T (2021) Congruency between two traditional and eDNA-based sampling methods in characterising taxonomic and trait-based structure of fish communities and community-environment relationships in lentic environment. Ecol Ind 129:107952
Darling JA, Jerde CL, Sepulveda AJ (2021) What do you mean by false positive? Environmental DNA 3:879–883
Deiner K, Fronhofer EA, Mächler E, Walser J-C, Altermatt F (2016) Environmental DNA reveals that rivers are conveyer belts of biodiversity information. Nat Commun 7:1–9
Dejean T, Valentini A, Duparc A, Pellier-Cuit S, Pompanon F, Taberlet P, Miaud C (2011) Persistence of environmental DNA in freshwater ecosystems. PLoS ONE 6:e23398
Erickson RA, Merkes CM, Mize EL (2019) Sampling designs for landscape-level eDNA monitoring programs. Integr Environ Assess Manag 15:760–771
Fediajevaite J, Priestley V, Arnold R, Savolainen V (2021) Meta-analysis shows that environmental DNA outperforms traditional surveys, but warrants better reporting standards. Ecol Evol 11:4803–4815
Ficetola GF, Pansu J, Bonin A, Coissac E, Giguet-Covex C, De Barba M, Gielly L, Lopes CM, Boyer F, Pompanon F (2015) Replication levels, false presences and the estimation of the presence/absence from eDNA metabarcoding data. Mol Ecol Resour 15:543–556
Friedrich T (2018) Danube sturgeons: past and future. Riverine Ecosyst Manag 8:507–518
Friedrich T, Pekarík L, Reinartz R, Ratschan C (2016) Restoration programs for the Sterlet (Acipenser ruthenus) in the upper and middle Danube. Danube News 33:4–5
Friedrich T, Gessner J, Reinartz R, Striebel B (2018) Pan-European action plan for sturgeons. Convention on the Conservation of European wildlife and natural habitats, Strasbourg, p 85
Friedrich T, Reinartz R, Gessner J (2019) Sturgeon re-introduction in the upper and middle Danube River Basin. J Appl Ichthyol 35:1059–1068
Guti G (2008) Past and present status of sturgeons in Hungary and problems involving their conservation. Large Rivers. https://doi.org/10.1127/lr/18/2008/61
Guy CS, Brown ML (2007) Analysis and interpretation of freshwater fisheries data. American Fisheries Society, Maryland
Hänfling B, Lawson Handley L, Read DS, Hahn C, Li J, Nichols P, Blackman RC, Oliver A, Winfield IJ (2016) Environmental DNA metabarcoding of lake fish communities reflects long-term data from established survey methods. Mol Ecol 25:3101–3119
Hill D, Fasham M, Tucker G, Shewry M, Shaw P (2005) Handbook of biodiversity methods: survey, evaluation and monitoring. Cambridge University Press
Holčík J (1989) The Freshwater Fishes of Europe: General Introduction to Fishes. Aula-Verlag, Acipenseriformes
ICPDR (2018) Sturgeon Strategy, ed. E.G.u.P.A.B.o.t.E.S.f.t.D. Region”
IUCN (2021) The IUCN Red List of Threatened Species
Jarić I, Gessner J, Solow AR (2016) Inferring functional extinction based on sighting records. Biol Cons 199:84–87
Jarić I, Riepe C, Gessner J (2018) Sturgeon and paddlefish life history and management: Experts’ knowledge and beliefs. J Appl Ichthyol 34:244–257
Jerde CL (2021) Can we manage fisheries with the inherent uncertainty from eDNA? J Fish Biol 98:341–353
Kubala M, Farský M, Krajč T, Pekárik L (2021) Bayesian modelling suggests that the sterlet (Acipenser ruthenus, Linnaeus 1758) population is ageing in the middle Danube River. Aquat Conserv Mar Freshwat Ecosyst 31:469–479
Lenhardt M, Jaric I, Kalauzi A, Cvijanovic G (2006) Assessment of extinction risk and reasons for decline in sturgeon. Biodivers Conserv 15:1967–1976
Luk’Yanenko V, Vasil’Ev A, Luk’Yanenko V, Khabarov M (1999) On the increasing threat of extermination of the unique Caspian sturgeon populations and the urgent measures required to save them. J Appl Ichthyol 15:99–102
Maasri A, Jähnig S, Adamescu M, Adrian R, Baigun C, Baird D, Batista-Morales A, Bonada N, Brown L, Cai Q (2021) A Global Agenda for Advancing Freshwater Biodiversity Research. Authorea Preprints
MacConaill LE, Burns RT, Nag A, Coleman HA, Slevin MK, Giorda K, Light M, Lai K, Jarosz M, McNeill MS (2018) Unique, dual-indexed sequencing adapters with UMIs effectively eliminate index cross-talk and significantly improve sensitivity of massively parallel sequencing. BMC Genomics 19:1–10
MacKenzie DI, Nichols JD, Sutton N, Kawanishi K, Bailey LL (2005) Improving inferences in population studies of rare species that are detected imperfectly. Ecology 86:1101–1113
Meulenbroek P, Hammerschmied U, Schmutz S, Weiss S, Schabuss M, Zornig H, Shumka S, Schiemer F (2020) Conservation requirements of European Eel (Anquilla anquilla) in a Balkan catchment. Sustainability 12:8535
Paraschiv M, Suciu R, Suciu M (2006) Present state of sturgeon stocks in the Lower Danube River, Romania, In Proceedings of the 36th International Conference of IAD, Austrian Committee Danube Research. pp. 4–8
Pfleger MO, Rider SJ, Johnston CE, Janosik AM (2016) Saving the doomed: Using eDNA to aid in detection of rare sturgeon for conservation (Acipenseridae). Global Ecol Conserv 8:99–107
Pont D, Rocle M, Valentini A, Civade R, Jean P, Maire A, Roset N, Schabuss M, Zornig H, Dejean T (2018) Environmental DNA reveals quantitative patterns of fish biodiversity in large rivers despite its downstream transportation. Sci Rep 8:1–13
Pont D, Keskin E, Meulenbroek P, Ünal EM, Bammer V, Dejean T, Er A, Erős T, Jean P, Lenhardt M (2021) Metabarcoding of fish eDNA samples. In: Igor Liška FW, Sengl M, Deutsch K, Slobodník J, Paunović M (eds) Joint danube survey4 - scientific report: a shared analysis of the Danube River. ICPDR Commission for the Protection of the Danube River, Vienna, p 565
Pont D, Meulenbroek P, Bammer V, Dejean T, Erős T, Jean P, Lenhardt M, Nagel C, Pekarik L, Schabuss M, Stoeckle B, Stoica E, Zornig H, Weigand A, Valentini A (2022) Quantitative monitoring of diverse fish communities on a large scale combining eDNA metabarcoding and qPCR. preprint
Rees HC, Maddison BC, Middleditch DJ, Patmore JR, Gough KC (2014) The detection of aquatic animal species using environmental DNA–a review of eDNA as a survey tool in ecology. J Appl Ecol 51:1450–1459
Reinartz R (2008) Artenhilfsprogramm Sterlet- Abschlussbericht 2004–2007. Landesfischereiverband Bayern, Münster
Reinartz R, Slavcheva P (2016) Saving sturgeons—A global report on their status and suggested conservation strategy. WWF, Vienna
Reinartz R, Lippold S, Lieckfeldt D, Ludwig A (2011) Population genetic analyses of Acipenser ruthenus as a prerequisite for the conservation of the uppermost Danube population. J Appl Ichthyol 27:477–483
Rourke ML, Fowler AM, Hughes JM, Broadhurst MK, DiBattista JD, Fielder S, Wilkes Walburn J, Furlan EM (2022) Environmental DNA (eDNA) as a tool for assessing fish biomass: a review of approaches and future considerations for resource surveys. Environmental DNA 4:9–33
Sandu C, Reinartz R, Bloesch J (2013) Sturgeon 2020”: A program for the protection and rehabilitation of Danube sturgeons. Danube Sturgeon Task Force (DSTF) & EU Strategy for the Danube River (EUSDR) Priority Area (PA)
Schnell IB, Bohmann K, Gilbert MTP (2015) Tag jumps illuminated–reducing sequence-to-sample misidentifications in metabarcoding studies. Mol Ecol Resour 15:1289–1303
Simonovic P, Budakov L, Nikolic V, Maric S (2005) Recent record of the ship sturgeon Acipenser nudiventris in the middle Danube (Serbia). Biologia 60:231–233
Sommerwerk N, Hein T, SchneiderJakoby M, Baumgartner C, Ostojić A, Paunović M, Bloesch J, Siber R, Tockner K (2009) The Danube river basin. In: Tockner K, Uehlinger U, Robinson CT (eds) Rivers of Europe. Elsevier, Amsterdam, pp 59–112
Stoeckle MY, Soboleva L, Charlop-Powers Z (2017) Aquatic environmental DNA detects seasonal fish abundance and habitat preference in an urban estuary. PLoS ONE 12:e0175186
Streibl D (1920) Über den Sterlet. Der Berufsfischer 2, Nr. 16
Szalóky Z, György ÁI, Tóth B, Sevcsik A, Specziár A, Csányi B, Szekeres J, Erős T (2014) Application of an electrified benthic frame trawl for sampling fish in a very large European river (the Danube River)–Is offshore monitoring necessary? Fish Res 151:12–19
Taberlet P, Coissac E, Hajibabaei M, Rieseberg LH (2012) Environmental DNA. Mol Ecol 21:1789–1793
Thomsen PF, Willerslev E (2015) Environmental DNA–An emerging tool in conservation for monitoring past and present biodiversity. Biol Cons 183:4–18
Thomsen PF, Kielgast J, Iversen LL, Møller PR, Rasmussen M, Willerslev E (2012) Detection of a diverse marine fish fauna using environmental DNA from seawater samples. PLoS ONE 7:e41732
Valentini A, Taberlet P, Miaud C, Civade R, Herder J, Thomsen PF, Bellemain E, Besnard A, Coissac E, Boyer F (2016) Next-generation monitoring of aquatic biodiversity using environmental DNA metabarcoding. Mol Ecol 25:929–942
Vassilev M, Pehlivanov L (2003) Structural changes of sturgeon catches in the Bulgarian Danube section. Acta Zoologica Bulgarica 55:97–102
Wiesner C, Schotzko N, Černý J, Guti G, Davideanu G, Jepsen N (2008) Technical report with results from the fish sampling and analyses from the Joint Danube Survey 2007
Xu N, Zhu B, Shi F, Shao K, Que Y, Li W, Li W, Jiao W, Tian H, Xu D (2018) Monitoring seasonal distribution of an endangered anadromous sturgeon in a large river using environmental DNA. Sci Nat 105:62
Yusishen ME, Eichorn F-C, Anderson WG, Docker MF (2020) Development of quantitative PCR assays for the detection and quantification of lake sturgeon (Acipenser fulvescens) environmental DNA. Conserv Genet Resour 12:17–19
Zajicek P, Wolter C (2018) The gain of additional sampling methods for the fish-based assessment of large rivers. Fish Res 197:15–24
Zauner G (1997) Acipenseriden in Österreich. Österreichs Fischerei 50:183–187
Acknowledgements
This work was supported by the Austrian Science Fund (FWF) project RIMECO (I 5006), the Austrian Federal Ministry for Digital and Economic Affairs, the National Foundation for Research, Technology and Development and the Christian Doppler Research Association, the Hungarian ANN-OTKA 135082 grant, the EU COST Action DNAqua-Net (CA15219), the INTERREG “MEASURES” program (DTP2-038-2.3), the Austrian Federal Ministry of Agriculture, Regions and Tourism (BMLRT), the LIFE-Programme of the European Union through the “LIFE-Sterlet” project and the ÖK-IAD (Österreichisches Komitee der Internationalen Arbeitsgemeinschaft Donauforschung). We would like to express our thanks to Jakob Neuburg, Daniel Trauner, Christoffer Nagel, Bernhard Stoeckle for their kind help in the field work and SPYGEN staff for their help in laboratory.
Funding
Open access funding provided by University of Natural Resources and Life Sciences Vienna (BOKU). This work was supported by the Austrian Science Fund (FWF) project RIMECO (I 5006), the Austrian Federal Ministry for Digital and Economic Affairs, the National Foundation for Research, Technology and Development and the Christian Doppler Research Association, the Hungarian ANN-OTKA 135082 grant, the EU COST Action DNAqua-Net (CA15219), the INTERREG “MEASURES” program (DTP2-038–2.3), the Austrian Federal Ministry of Agriculture, Regions and Tourism (BMLRT), the LIFE-Programme of the European Union through the “LIFE-Sterlet” project, the ÖK-IAD (Österreichisches Komitee der Internationalen Arbeitsgemeinschaft Donauforschung), and the Ministry of Education, Science and Technological Development of the Republic of Serbia (Projects 451-03-68/2022-14/200053 and 451-03-68/2022-14/200007).
Author information
Authors and Affiliations
Contributions
MP, PD: Conceptualization, MP, VA. PD: Formal analysis, MP, SM, PD, ZH, LM, LP, FT: Investigation, VA, PJ, TD, PD: Methodology, MP, HT, PD: Project administration, MP: Writing—original draft, Writing, All authors: review & editing.
Corresponding author
Ethics declarations
Conflict of interest
‘teleo’ primers and the use of the amplified fragment for identifying fish species from environmental samples are patented by the CNRS and the Université Grenoble Alpes. This patent only restricts commercial applications and has no implications for the use of this method by academic researchers. SPYGEN owns a license for this patent. Three coauthers are research scientists at a private company specialising in the use of eDNA for species detection.
Additional information
Communicated by Paul Humphries.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Meulenbroek, P., Hein, T., Friedrich, T. et al. Sturgeons in large rivers: detecting the near-extinct needles in a haystack via eDNA metabarcoding from water samples. Biodivers Conserv 31, 2817–2832 (2022). https://doi.org/10.1007/s10531-022-02459-w
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10531-022-02459-w