Abstract
Harmonia axyridis is an invasive beetle (Coleoptera: Coccinellidae), native to Asia, that has established on four continents beyond its native range and was first observed in South Africa in 2001. Using microsatellite data and a comprehensive geographic coverage of international (17 locations) and South African (14 locations) beetle samples, we inferred the source of colonists invading South Africa and the beetle’s movement within the country. Genetic structure analyses suggest that this species’ populations in South Africa are derived from admixture between beetles originating from eastern North America (which acted as a bridgehead population) and another population not directly sampled in our study. Genetic admixture prior its invasion into South Africa, along with the possibility of multiple introductions from the bridgehead population, may explain the high genetic diversity estimates for South African H. axyridis (HE = 0.582–0.625) compared to beetles from the native range (HE = 0.440–0.636). Harmonia axyridis appear to be frequently exchanging genetic material across South Africa, with no clear genetic structure between sampled locations. However, evidence of weak isolation-by-distance within South Africa suggests that beetles are dispersing less frequently between locations that are furthest apart. This study supports previous findings on global invasion pathways in this species but provides new insights in the context of the invasion in South Africa. We highlight how mixing of distinct lineages from divergent origins prior to the invasion into South Africa has augmented genetic diversity in the region. The high dispersal rates and large effective population sizes inferred from genetic markers suggest that slowing the spread or reducing population abundances of the species in South Africa will be challenging without an integrated, multi-faceted management approach.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Dispersal pathways and demographic characteristics are important factors affecting the introduction and spread of invasive species (Kenis et al. 2007; Caley et al. 2015). Most invasive species are introduced unintentionally, and as a result go undetected for some time (Lockwood et al. 2005; Hill et al. 2016; Roques et al. 2016). In some cases, intentionally introduced species, like those reared for biological control of pest species in agricultural systems, can accidentally spread to novel environments where it was not intended for release (Pratt and Center 2012). This can be a cause for concern because species introduced intentionally for cultivation or biological control are given special attention to promote persistence and to overcome barriers to introduction (Wilson et al. 2009). Non-native species whose establishment was facilitated by human activities may have adaptive advantages when accidentally introduced elsewhere (e.g., Anthropogenically Induced Adaptations to Invade, or AIAI; Hufbauer et al. 2012). Therefore, investigating the routes of introduction is a critical step toward mitigating against the arrival and establishment of potential future invaders (Estoup and Guillemaud 2010) and to elucidate the processes that facilitate ongoing invasions (Wilson et al. 2009).
Invasive populations very often show evidence of multiple introductions and/or high propagule pressure and can be accompanied by admixture events between two or more genetically divergent populations (Garnas et al. 2016; van Boheemen et al. 2017). The merging of previously isolated lineages can result in admixed genotypes that are not typically found in the native range (Keller and Taylor 2008; van Boheemen et al. 2017). Genetic admixture can further influence the adaptive ability of introduced individuals by elevating additive genetic variation and/or creating novel gene combinations upon which selection can act. Such mixing can also ameliorate the negative effects associated with genetic bottlenecks and inbreeding and aid adaptation to novel conditions (Estoup and Guillemaud 2010; Lombaert et al. 2010; Bock et al. 2015). However, gene flow between lineages with distinct evolutionary histories can also lead to genomic incompatibilities or compromise local adaptation (Garnas et al. 2016). Sufficient additive genetic variation has often been cited as a key factor influencing the ability of invasive species to cope with heterogeneous and/or novel environmental conditions, though others have recognized an important role for phenotypic plasticity (Rius and Darling 2014). The success of numerous invaders with limited genetic diversity, including clonal forms, attests to the fact that genetic variation is not universally required for ecological success, at least in the short term (Garnas et al. 2018). Still, high propagule pressure, either at the outset of establishment or via multiple introduction events, generally correlates with population genetic diversity and positively relates to introduced species’ success (Wilson et al. 2009). Within the last century, several countries have increased border security and quarantine measures to combat the spread of invasive species, though rates of establishment have not dramatically changed (Seebens et al. 2017). Non-native insects and other invertebrates are particularly well-represented among invasive taxa, likely due to their unique capacity to disperse over large distances undetected as contaminants and stowaways on global trade and transport channels (Roques et al. 2016; Seebens et al. 2017; Van Wilgen et al. 2020).
Harmonia axyridis (Pallas 1773) (Coleoptera: Coccinellidae), commonly known as the harlequin ladybird, is a beetle native to Asia (Koch 2003; Brown et al. 2011). It gained attention for voraciously feeding on aphids and effectively suppressing their populations, particularly in greenhouse or nursery environments (Koch 2003). As a consequence of its success as a commercial biocontrol agent, this species was repeatedly introduced outside of its native range (Koch 2003). It was first intentionally released into Europe (1916) and North America (1964) (Iablokoff-Khnzorian 1982; Chapin and Brou 1991), followed by recurrent intentional introductions into parts of South America between 1986 in 1998 and at least three countries in Africa: South Africa (1980), Tunisia (1990) and Egypt (before 2000), although it only established in the latter region (Poutsma et al. 2007; Brown et al. 2011; Roy et al. 2016). After decades of introducing H. axyridis to manage invertebrate pest species, introduced populations began gradually expanding their ranges (Poutsma et al. 2007), and by the late 1900s the species was recorded in regions where it had not been introduced and/or where it had putatively failed to establish (Brown et al. 2011; Roy et al. 2016). It was first observed in the wild and recognised as an invasive species in North America in 1988 (Oregon, USA) and 1991 (Louisiana, USA) (Brown et al. 2011). Several years later, wild populations were observed in Europe (Belgium), South America (Argentina), and Africa in 2001 (South Africa) (Stals and Prinsloo 2007; Brown et al. 2011). A landmark study of its worldwide population genetics suggested that beetles from two distinct genetic groups in the native range in Asia were introduced to eastern North America where an admixture event occurred between two genetically distinct groups (Lombaert et al. 2014). Individuals resulting from this admixture event in eastern North America then acted as the source of colonists invading South America, South Africa, and parts of Europe. Lombaert et al. (2010) described this phenomenon as a bridgehead effect, whereby individuals originating from a single region acted as the source of colonists invading multiple new territories. Harmonia axyridis has since been recorded in at least 59 countries on four continents outside of its native range (Camacho-Cervantes et al. 2017) where its presence negatively influences native invertebrate species, including native ladybirds (Michaud 2002; Brown et al. 2011; Roy et al. 2016). Despite its primary role as a natural enemy, H. axyridis is also recognised as an agricultural pest, specifically in vineyards where beetles are so abundant that they can contaminate grape harvests and reduce wine quality (Pickering et al. 2004; Galvan et al. 2008). Further, these beetles are considered a nuisance when they invade homes, particularly in the search for protected overwintering sites, or due to their moderately painful bite. In some cases, interaction with beetles can cause allergic reactions in people (Roy et al. 2016; Camacho-Cervantes et al. 2017).
In South Africa, the first observations of H. axyridis were made in Stellenbosch and Cape Town, Western Cape Province in 2001 (Stals 2010). This comes two decades after a purportedly failed attempt to introduce the beetle was made during the 1980s to manage the black pine aphid, Cinara cronarti, in the Mpumalanga Province (Roy et al. 2016), approximately 1,500 km northeast of the Western Cape. Since 2001, this beetle has been detected in most regions of the country and in two neighbouring countries Lesotho and eSwatini (Stals 2010; Nedvěd and Háva 2016). However, several key details about its invasion history and movement within South Africa remain unknown. Population genetics provide methods of determining population demographic history (Chapuis and Estoup 2007) using genetic markers such as microsatellites. Previous genetic studies have used microsatellite data to make inferences about the worldwide invasion history of H. axyridis (Lombaert et al. 2010, 2011, 2014; Facon et al. 2011). Even though the studies by Lombaert et al. (2011, 2014) included a large set of sampling localities across most of the species’ current distribution, including its native and invaded ranges, sampling in South Africa in 2008 was limited, comprising only four sampling sites across the country in the Western Cape (Somerset West), Gauteng (Benoni), KwaZulu Natal (Nottingham Road), and the Free State (Bethlehem) Provinces.
In this study, we investigate the global genetic structure of H. axyridis with a focus on South African populations and determine the likely route(s) of invasion into the country and the movement within. To achieve this, we included a comprehensive geographic coverage of beetles and a systematic sampling of multiple South African locations. We examine how our results fit into the global invasion network proposed by Lombaert et al. (2014). This approach can provide insights on both fine- and broad-scale movements of H. axyridis in South Africa and worldwide. Furthermore, we sought to compare our findings with previous population genetic studies conducted nearly ten years prior (Lombaert et al. 2010, 2011, 2014; Facon et al. 2011) both to confirm and to assess the temporal stability of the invasion pathways and patterns of spread of H. axyridis. We expect that as an invasive species becomes increasingly well-studied and public awareness grows, and as border biosecurity simultaneously tightens, secondary invasion from elsewhere in the global range would become less likely, and that genetic diversity would stabilize or decline (i.e., as populations adapt to local conditions). Alternatively, ongoing cryptic invasions and/or continued rapid population growth would lead to increased genetic diversity over time. Detecting such patterns requires repeated sampling and analyses (i.e., 2010 to current) and has not been yet undertaken for H. axyridis in South Africa.
Materials and methods
Sampling
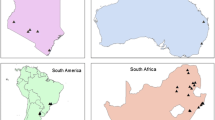
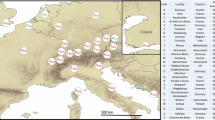
Harmonia axyridis samples were collected between 2016 and 2020 with the help of a citizen science campaign launched in 2016 by S. Clusella-Trullas through the Centre for Invasion Biology (CIB) that encouraged citizens to submit samples of H. axyridis collected throughout the country to Stellenbosch University. In South Africa, 15–35 individuals from each of the 14 sampling locations were selected for genetic analyses (Fig. 1, Table 1). Eight additional sampling locations were included from the native region in Asia and nine from introduced populations in North America, South America, and Europe (Fig. 1, Table 1). We also obtained samples of an invasive population of H. axyridis recently detected in northern Greece that had not been included in previous genetic studies of this beetle. This Greek population was detected in 2017 (Ceryngier and Romanowski 2017) in a region that appeared to be ‘resistant’ to the establishment of H. axyridis (Roy et al. 2016).
Sampling locations of Harmonia axyridis across native and invaded ranges showing more focused sampling in South Africa. Grey and red dots are invasive and native populations, respectively. Sample codes are described in Table 1
DNA extraction and microsatellite genotyping
Genomic DNA was extracted from whole specimens using a QIAGEN DNeasy® blood and tissue kit (Qiagen Inc., Hilden, Germany), following the manufacturer’s instructions. All individuals were then genotyped at 15 microsatellite makers developed in a previous study (Loiseau et al. 2009; Online Resource 1). To allow for multiplexing, the forward primers were labelled at the 5’ end and pooled in three multiplex sets with one set of eight, one of four, and one of three primers to yield unique index combinations (the latter two sets were pooled before genotyping) (Online Resource 1). A QIAGEN multiplex PCR kit (Qiagen Inc., Inc., Hilden, Germany) was used to perform the amplification process following the PCR conditions described in Loiseau et al.2009. To ensure consistent plate reading we included a negative and positive control in each plate. Samples were genotyped using an ABI 3130 Automated sequencer (Applied Biosystems, Foster City, California, USA). Samples which were unsuccessfully amplified after two or three independent runs were considered non-amplifiable and subsequently not included in the dataset. Samples which showed successful amplification after a second or third run were scored again along with the completed set of individuals included in our study to ensure consistent scoring of alleles. Alleles were scored using Applied Biosystems™ Microsatellite Analysis Software™ CE Fragment Sizing.
Microsatellite analyses
Sample variability
GENEPOP v4.01 (Raymond and Rousset, 1995; Rousset, 2008) was used to test deviations from Hardy–Weinberg equilibrium (HWE) and linkage disequilibrium (LD) using 1000 iterations per batch. Genetic variation was investigated by calculating basic descriptive statistics including number of alleles (NA), observed heterozygosity (HO) and expected heterozygosity (HE) (GENETIX 4.50.2, Belkhir et al. 2004). FSTAT v. 2.9.4 (Goudet 2001) was used to calculate allelic richness (AR) and inbreeding coefficient (FIS, Table 1). The frequency of null alleles (AN) was estimated using FreeNA v.1.0 (Chapuis and Estoup 2007), and the number of private alleles (NP) was calculated using GenAlEx v. 6.5 (Peakall and Smouse 2006, 2012). Furthermore, we used one-sided Wilcoxon sign rank tests to detect if any of the sample locations experienced a genetic bottleneck. After a genetic bottleneck, populations are predicted to lose allelic diversity faster than heterozygosity (Luikart and Cornuet 1998). Therefore, an excess in heterozygosity can be used as an indicator of a recent genetic bottleneck. We used BOTTLENECK v1.2.0.2 (Cornuet and Luikart 1996) implementing two models: two-phase model of mutation (TPM) and the stepwise mutation model (SMM) to determine if populations displayed deviations from expected heterozygosity.
Population structure
To determine the degree of genetic differentiation between all sampling locations, pairwise FST values were calculated according to the method described by Weir and Cockerham (1984) using HIERFSTAT v.0.5–7 (Goudet and Jombart 2015) in R version 4.0.4 (2021–03-22). Overall FST was calculated using FreeNA v1.0, both including and excluding null alleles and using 10 000 replicates (Chapuis and Estoup 2007). To further describe the genetic relationships between the sampled populations, we used the R package ADEGENET v.2.1.3 (Jombart and Ahmed 2011) to conduct a Correspondence Analysis (CoA) and used the PC values in a Discriminate Analysis of Principal Components (DAPC), which utilizes a multivariate approach to identify genetic clusters of individuals. We inferred the number of clusters by using the “find.clusters” function and used BIC to determine the optimal number of clusters. This was done for the global dataset, as well as for South African sampling locations independently.
We then used POPTREEW (Takezaki et al. 2014), an online software [http://www.med.kagawa-u.ac.jp/~genomelb/takezaki/poptreew/index.html], to construct an unrooted Neighbour-Joining population tree for H. axyridis based on shared alleles (DS; Bowcock et al. 1994). We used Figtree v.1.4.4 (Rambaut 2010) to visualise the Neighbour-Joining tree. Statistical support was assessed for each branch using non-parametric bootstrapping of 10 000 replicates. ARLEQUIN v3.5.2.2 (Excoffier and Lischer 2010) was used to investigate the isolation-by-distance hypothesis (Wright 1946) of H. axyridis within South Africa by conducting separate Mantel’s tests for each of the four geographic groups (Americas, Europe, Asia, and South Africa). Furthermore, separate Mantel’s tests were conducted to investigate isolation-by-distance between populations in South Africa that were closest to two of South Africa’s seaport harbours: the Port of Durban and the Port of Cape Town, and populations further inland. Impendile (S-IMP) and Marwaqa Nature Reserve (S-MWA) were used to represent populations closest to the Port of Durban and the populations from Hout Bay (S-HBY) and Stellenbosch (S-STB) were used to represent populations closest to the Port of Cape Town.
Population genetic structure was assessed using STRUCTURE 2.3.4 (Pritchard et al. 2000; Falush et al. 2003), a programme which implements Bayesian clustering methods to define possible genetic units, or clusters (K), based on the multilocus genotypes of individuals without providing any prior spatial information to the software. We employed an admixture ancestry model with correlated allele frequencies which allows allele frequencies within different populations to be similar due to shared ancestry or migration between populations. We carried out ten independent replicated runs for each value of K set between 1 and 29 (where 29 is the total number of locations sampled) using a burn-in set to 100 000 followed by 1 000 000 Markov Chain Monte Carlo (MCMC) permutations. To identify the optimal number of clusters (i.e., the most probable value of K) we used the delta K (ΔK) as described by Evanno et al. (2005). Delta K is based on the second order rate of log probability change of successive K values (Ln P(X/Y)), which provides an estimation of the most-true value of K (Evanno et al. 2005). These calculations were implemented using the programme CLUMPAK v 1.1 (Kopelman et al 2015). The ΔK method described by Evanno et al. (2005) is limited by the fact that it only detects the uppermost limit of genetic structure while several hierarchical genetic structure levels may exist within the dataset. To account for this, we independently reanalysed all major clusters until no genetic structure was identified in the group following the method described in Karsten et al. (2015) in addition to reanalysing subsets of data according to geographic location (South Africa, Americas, Europe, Asia) following the methods described in Javal et al. (2018) to investigate further sub-structuring in the data.
Finally, we conducted an Analysis of Molecular Variance (AMOVA) for five different hypothetical population structure scenarios using the programme ARLEQUIN v3.5.2.2 (Excoffier and Lischer 2010). The population structure scenarios were based on partitioned sampled populations according to geographic subdivision, co-ancestry results from STRUCTURE, and the genetic relationships inferred from the neighbour-joining tree.
Colonisation history
To infer the colonisation history of H. axyridis, we tested different hypothetical scenarios describing the invasion history of each genetic group identified from population structure analyses using the Approximate Bayesian Computation (ABC) method in the programme DIYABC v2.1.0 (Cornuet et al. 2014). ABC allows the comparison of intricate population histories using simulations based on summary statistics and provides support estimates for the most likely scenario choice including posterior probability estimates with 95% confidence intervals (Guillemaud et al. 2010; Cornuet et al. 2014). An advantage of the ABC method is that it has the ability to simulate unsampled or “ghost” populations that may have contributed to the invasion history of populations, but that were not sampled (Lombaert et al. 2011). Unsampled populations can then be included in the hypothetical scenarios tested for each geographic group. The DIYABC programme will provide support estimates to suggest if it was more/less likely that a “ghost” population was involved in the invasion history of the population(s) being tested.
This study included many sampling locations and the scenarios tested were narrowed down by including historical records (or information of first introduction), results from our cluster analyses (STRUCTURE) and to avoid violating the coalescent model assumptions implemented in the DIYABC programme, we selected a single sample location to represent each genetic cluster (Online Resource 2). We tested 99 scenarios considering six broad hypotheses for the different geographic areas invaded by H. axyridis (Online Resource 2).
The first set of competing scenarios we tested for eastern North America (1988) included introductions from populations from the native region and genetic admixture between those populations. Invasive populations of H. axyridis were next observed in west North America in 1991. South America, Europe and South Africa had its first observation records of H. axyridis in the wild in the same year (2001). For South America we tested two hypothetical scenarios for this region: the first scenario included a direct introduction from eastern North America, and the second included the same introduction scenario as we found for eastern North America. Comparing these two scenarios could discern whether it is more likely that beetles were introduced directly from eastern North America, or if the introduction of H. axyridis into South America followed the same introduction route as it did for eastern North America. Scenarios for the invasion of Europe and South Africa included introduction scenarios from each genetic group identified in the population structure analyses (excluding Greece), and admixture scenarios between each of those populations. We also included unsampled, or “ghost”, populations in these tests. Finally, the most recent first observations of H. axyridis amongst the populations sampled for this study was in Greece in 2017. Similar to the scenarios tested for Europe and South Africa, we included all plausible introduction scenarios including introductions from populations from each genetic group identified in the population structure analyses and admixture scenarios between each of those populations. We also included unsampled populations in these tests.
DIYABC measures the timing of events for the historical scenarios using the number of generations the study organism can support within a year, we therefore expressed all times assuming 2.5 generations per year (Osawa 2000; Lombaert et al. 2011, Online Resource 3). We simulated 1 000 000 computations per scenario tested. The posterior probability for each computed scenario was estimated with 95% confidence intervals (CI), including a logistic regression. The most likely scenario was chosen based on the highest posterior probability value with a non-overlapping 95% confidence interval.
Results
Genetic diversity
Overall, 83 of 403 population/locus combinations showed significant deviations from HWE; no significant linkage disequilibrium was found for any population included in our study. The mean null allele frequency (NA) in all populations ranged between 0.00 (Irkutsk, Russia) and 0.07 (Thessaloniki, Greece). South African populations showed relatively high levels of genetic diversity relative to the native range, with mean expected heterozygosity (HE) ranging between 0.58 (Sani Pass) and 0.625 (Sandfontein), number of alleles (NA) ranging between 4.87 (Silverdale) and 6.07 (Sandfontein), and allelic richness (AR) ranging between 3.69 (Sani Pass) and 4.06 (Stellenbosch) (Table 1). Descriptive statistics for the North and South American (HE: 0.61–0.68), and European (HE: 0.60–0.65) introduced populations were comparable to those from South Africa, also displaying high levels of genetic diversity compared to those from the native range (0.44–0.64) (Table 1). In South Africa, four populations had a few private alleles (0.13 in Bredasdorp and 0.07 in De Zalze, Goedehoop, and Sandfontein, respectively; Table 1). Vancouver (Canada) showed the highest number of private alleles (0.47) while several locations in the native region (Wuhan, Irkutsk, Tomsk) had no private alleles. Recent bottlenecks, identified by significant excess heterozygosity (p < 0.05), were detected in all but two South African populations (De Zalze Golf Estate and George) (Online Resource 4). Recent bottlenecks were also detected in Europe (Tours, France), South America (Santiago, Chile), and two populations in the native range (Changchun (China) and Irkutsk (Russia)) (Online Resource 4). Furthermore, populations in Europe (Tervuren), North America (Vancouver) and four populations in the native range (Khakassia, Krasnoyarsk, Primorsky Krai, and Tomsk) showed significant heterozygosity deficit, indicative of an expanding population (Online Resource 4).
Population structure
Differentiation of allele frequencies between each population was measured using FST (Online Resource 5). Pairwise FST values ranged between − 0.008 (Goedehoop and Silverdale) and 0.218 (Changchun and Irkutsk) with only a small percentage of pairwise comparisons (5.98%) showing no significant difference indicative of genetic differentiation (Online Resource 5). This percentage was mostly made up of within-southern Africa comparisons and between five Asian populations in the native region (Kyoto and Tomsk; Khakassia and Krasnoyarsk; Primorsky Krai and Tomsk). Overall FST values before and after excluding null alleles correction (0.0637 and 0.0635 respectively) indicated significant population differentiation.
The unrooted NJ tree based on shared alleles (DS; Bowcock et al. 1994) identified four genetic clusters that largely reflected country/continent of collection: South Africa, Europe, Americas (excluding Vancouver) and Asia (Fig. 2). The Asian group contained all native H. axyridis populations as well as the non-native western North American population from Vancouver, Canada (Fig. 2). The distinction between the genetically related clusters was less clear from the DAPC output based on the lowest BIC value (Fig. 3). One major cluster included all South African populations, while another cluster included populations from the rest of the world (Fig. 3A). A single location, Irkutsk (Russia), was the only population that did not fall within either of these two major clusters. Irkutsk displayed evidence of a genetic bottleneck, which may reflect an ecological or physical event in the environment of H. axyridis in this region resulting in a population contraction (Ramakrishnan et al. 2005). To investigate further substructure among South African populations, we used Discriminant Analysis of Principle Components (DAPC) including only South African populations which showed virtually no significant pattern of genetic differentiation among sample localities. One exception was Bredasdorp, whose samples lay outside the main cluster (Fig. 3B). Results from the STRUCTURE clustering analyses indicated an optimal number of K = 2 (Fig. 4). This corresponded with the results from the DAPC of the global dataset where all Southern African populations belonged to a single genetic cluster, while the rest of the sampled populations fell outside of this group (Fig. 3A). Co-ancestry results from the K = 2 output suggests Minnesota and New Hampshire (east North America), Limache and Santiago (South America), and Thessaloniki (Greece) are more genetically similar to the South African populations than to the other sampled populations included in our study (Fig. 4). We investigated further substructure within each group of genetically similar populations. Within the group containing populations from South Africa, the Americas, and Greece, STRUCTURE clustering analyses indicated an optimal number of K = 3 (Fig. 4). Populations from South Africa clustered together, populations from eastern North America and South America clustered together, while the population from Greece belonged to its own distinct cluster. Population structure analyses of the two genetically distinct groups (South Africa and the Americas) revealed no further substructure (Fig. 4). We conducted the same substructure investigation for the populations collected from Belgium, Czech Republic, and France (Europe), Vancouver (west North America), and China, Japan, and Russia (Asia) that were found to be genetically similar. STRUCTURE clustering analyses indicated an optimal number of K = 4 for this group (Fig. 4). All of the European populations clustered together, while three genetically distinct groups were revealed made up of Asian populations and the west North American population. Here, they are identified as: Asia 1—made up of populations from Japan and Russia, and west North America; Asia 2—made up of populations from China and Russia, and Asia 3—made up of a population from China. Population structure was analysed a final time for each of the clusters (Fig. 4). No further substructure was revealed within each of the groups except the populations from Asia 2 which seemed to indicate that the two populations (Wuhan (China), and Irkutsk (Russia)) could be separated further into two more sub-groups. However, we still recognised it as a single group in this study because all other STRUCTURE genetic structure analyses (including STRUCTURE analyses according to geographic group, Online Resource 6) and neighbour-joining tree suggest that it is a single genetic group. Based on the STRUCTURE results after investigating further substructure within each subset of data, and investigating further substructure based on geographic location (Online Resource 6), we reached an optimal number of clusters at K = 7 (Figs. 4 and 5). This result matched closely with the Neighbour-Joining tree where South Africa, Europe, and the Americas (excluding Vancouver) belonged to three distinct genetic groups, and populations in western Asia (Russia: KRS, KHA, TOM) are more genetically similar to one another than they are to those further east (Fig. 2). However, results from the STRUCTURE analyses and DAPC in our study did not find a clear genetic difference between sample locations from eastern and western Asia, instead, sample locations furthest east and furthest west belong to the same genetic clusters, with two clusters falling in-between (Figs. 4 and 5). Moreover, significant differences were found among all groups in each scenario tested using an analysis of molecular variance (AMOVA) (Online Resource 8). Mantel’s tests for isolation-by-distance patterns showed no significant results for the native region (r = − 0.260; p = 0.96) and the European (r = 0.53; p = 0.14) and American (r = 0.43; p = 0.35) introduced ranges. However, a significant isolation-by-distance pattern was found in South Africa (r = 0.34; p = 0.01). Separate Mantel’s tests were conducted for grouped sampling localities in southwestern (r = 0.94, p = 0.26) and northeastern South Africa (r = 0.15, p = 0.9). These tests indicate no evidence of isolation-by-distance among sample localities within these regions. Finally, separate Mantel’s tests were conducted to investigate isolation-by-distance between South African populations closest to seaport harbours in Durban and Cape Town, and those further inland. The Mantel’s tests showed no significant IBD, indicating that distance from these seaport harbours do not influence genetic similarity between locations at the coast and further from the coast (Online Resource 9).
Unrooted Neighbour-Joining tree for genetic distance of H. axyridis, based on shared alleles of microsatellite markers (DS; Bowcock et al. 1994). Only bootstrap values above 70% after 10 000 replicates are shown at the nodes of branches. Colours demarcating groups of closely related populations from South Africa (yellow), Americas (red) and Europe (green) and a cluster including all native populations, and the invasive population in Vancouver (blue) correspond to clusters identified in STRUCTURE analyses. Sample codes are described in full in Table 1
Results of 1st and 2nd axes from the Discriminant Analyses of Principal Components (DAPC). Graphics as default from R package: Adegenet v.2.1.3. A First two principal components identifying clusters of genetically related Harmonia axyridis individuals for all populations sampled in our study (South Africa, Europe, North America, South America and Asia). B First two principal components identifying clusters of genetically related H. axyridis individuals sampled only from within South Africa. Each point represents a sampled individual and is coloured according to the population it belongs to. Clusters (ellipses) are drawn based on the lowest BIC value
Hierarchal coancestry assignment results of the global dataset of Harmonia axyiridis beetles from the programme STRUCTURE for optimal clusters according to the delta K (ΔK) method as described by Evanno et al. (2005). Structure was re-run for all major clusters until no genetic structure was identified within the group. Even though “ASIA 2 K = 2” illustrates that the group should be separated further into two more sub-groups. We still recognised it as a single group in this study because all other STRUCTURE genetic structure analyses and neighbour-joining tree suggest that it is a single genetic group. Each bar represents an individual and are grouped by geographic location in the order it appears in Table 1. (ENA = eastern North America (Minnesota, New Hampshire), WNA = western North America (Vancouver), SA = South America (Lim ache, Santiago), GR = Greece (Thessaloniki); Europe (Czech Republic, Belgium, France), A1 = Asia 1 (Vancouver, Russia, Japan), A2 = Asia 2 (Russia, China), A3 = Asia 3 (China))
Genetic clustering of international sampled populations of Harmonia axyridis. Sample location points are coloured according to the genetic cluster the population belongs to according to results from the programme STRUTURE for K = 7 (Fig. 4). The genetic clusters are as follows: Yellow = South Africa; Red = Europe (TER, RAK, TOU); Dark Green = Americas (MIN, HAM, LIM, SNT); Light Green = Greece (THS); Purple = Asia 1 (TOM, KRS, KHA, TOM, PRM, KYO, VAN); Pink = Asia 2 (IRK, WUH); Blue = Asia 3 (CHN). Sample codes are described in full in Table 1
Colonisation history
To investigate the global colonisation history of H. axyridis, competing scenarios were tested using Approximate Bayesian Computation in DIYABC for five genetic/geographic groups outside of the native range as identified from the population structure analyses (STRUCTURE) and utilising historical information (records of first introduction). These regions included eastern North America, South America, Europe (excluding Greece), South Africa and Greece (Figs. 5 and 6, Online Resource 2). Posterior probability estimates (with 95% confidence intervals) indicate that the most likely invasion scenario of H. axyridis into eastern North America involved genetic admixture between two distinct genetic groups from the native range, Asia 1 and Asia 2 (0.40 [0.40, 0.41]). The most favoured scenario had eastern North America as the sole source of the invasion of South America (0.99 [0.99, 1.00]). Posterior probability estimates indicated that beetles sampled from Europe showed admixture between beetles originating from western North America (Vancouver) and an unsampled population (0.72 [0.51, 0.93]). The South African invasion showed evidence of admixture between beetles originating from eastern North America and an unsampled population (0.66 [0.52, 0.81]). Finally, the Greek population was derived from genetic admixture between beetles originating from eastern North America and elsewhere in Europe (0.88 [0.83, 0.92]) (Online resource 2).
The most likely invasion route of Harmonia axyridis according to the STRUCTURE (Fig. 4, Fig. 5) and Approximate Bayesian Computation (ABC) analyses (Online Resource 2) conducted in our study. The scenario with the highest posterior probability estimates for each of the five introduced populations tested in the DIYABC software is listed in the table on the right. Each circle represents a sampled population included in our study and are coloured according to the genetic cluster it belongs to according to the STRUTURE analyses (RSA = South Africa, ENA = eastern North America (Minnesota, New Hampshire), WNA = western North America (Vancouver), SA = South America (Limache, Santiago), GR = Greece (Thessaloniki); EU = Europe (Czech Republic, Belgium, France), A1 = Asia 1 (Russia, Japan), A2 = Asia 2 (Russia, China), A3 = Asia 3 (China)) (Fig. 4, Fig. 5). Arrows indicate the most likely introduction scenario based on the ABC results from our study using the programme DIYABC v2.1.0 (Cornuet et al. 2014) (Online Resource 2). Dates indicate earliest observation of H. axyridis based on historical records and stars indicate that genetic admixture occurred between two populations. In the cases where genetic admixture occurred between a sampled population and an unsampled population, it is indicated in bold writing. The position of the “unsampled population” label on the map is arbitrary because we cannot know the origin of the unsampled population simulated by the DIYABC programme
Discussion
Population structure
Population structure analyses identified two main genetic groups within the global sampling of H. axyridis: South Africa grouped with eastern North America, South America, and Greece, while the sample populations from western North America, Europe, and Asia were more genetically related to each other. The two groups were then further subdivided into seven distinct genetic clusters: (1) eastern North America and South America; (2) Europe (all locations excluding Greece); (3) Greece; (4) South Africa, and three distinct groups within the native range (5) Asia 1 (Russia and Japan. This genetic cluster also included Vancouver, Canada), (6) Asia 2 (China and Russia), and (7) Asia 3 (China).
The lack of clear population genetic structure within South Africa is indicative of rapid spread and/or high gene flow across the country. Harmonia axyridis is known to disperse over several kilometres each year in search of overwintering sites (Raak-van den Berg et al. 2012), and it has been detected travelling at 60 km/hour, 1100 m above ground level using vertically-facing entomological radar (Jeffries et al. 2013). Additionally, mark-release field trials demonstrated H. axyridis beetles had an average flight distance of 431 m and 396 m for males and females respectively within approximately one month (Seko et al. 2008). Long-distance dispersal within South Africa is most likely facilitated by human-mediated processes. In South Africa, H. axyridis beetles have been observed on flowers of garden plants and ornamental conifers (Roy et al. 2016) and may be transported on ornamental plants from nurseries moving across the country (Roy and Brown 2015). In parts of Europe H. axyridis has been transported on produce (fruit, vegetables, flowers) and as stowaways on trains and motor vehicles (Brown et al. 2011). People travelling or transporting goods (harvested grapes and citrus) may be inadvertently facilitating the long-distance dispersal of this beetle across South Africa, such as is thought to be the case in the other important invaders (i.e., fruit flies within the Tephritidae; Karsten et al. 2015, 2018). This could also be an example of “stratified dispersal”: a combination of short- and long-distance dispersal that promotes genetic diversity across the landscape and could be indicative of an expanding population (Gilbert et al. 2004; Ciosi et al. 2011). However, evidence of moderate isolation-by-distance (Wright 1946) suggests that dispersal among regions furthest apart is probably quite rare. Isolation by distance occurs when gene flow declines with geographic distance (Wright 1946). Therefore, beetles within South Africa are dispersing less frequently between populations that are furthest apart. Furthermore, results from the DAPC including only South African samples indicate that the population sampled from Bredasdorp (Western Cape Province) differs genetically from the other samples included in our study. This difference was not seen in any of the other population structure analyses, including STRUCTURE and Neighbour-Joining algorithms. Why the population sampled from Bredasdorp differs from the rest of the H. axyridis populations in the DAPC is not currently known.
The lack of population genetic structure of H. axyridis within South Africa (again, with the exception of Bredasdorp) is consistent with earlier findings of a single genetic cluster within the country (Lombaert et al. 2010; 2011, 2014). The 2014 study included only four sampling locations from South Africa which also predate ours by a decade, relatively early on in the invasion, first detected in 2001. It is perhaps not surprising that significant population structure has not developed since then, especially in light of sustained gene flow as evidenced by the moderate strong IBD. Our results do diverge to some degree however with previous reconstructions of worldwide population structure, particularly in the native range and within Europe. For example, Lombaert et al. (2014) identified two genetic clusters in the native region: one in western Asia and a separate distinct cluster in eastern Asia. Our Neighbour-Joining tree suggests populations in western Asia (Russia: KRS, KHA, TOM) are more genetically similar to one another than they are to those further east. However, results from the STRUCTURE analyses and DAPC in our study did not find a clear genetic difference between sample locations from across Asia. Instead, STRUCTURE analyses show sample locations furthest east and furthest west belong to the same genetic cluster, with two clusters falling in-between. Additionally, population structure analyses from our study identified two genetic clusters in Europe, compared to the four genetic clusters previously identified. This is despite three of the four sample locations included in our study (Belgium, France, Czech Republic) – which belong to a single distinct genetic cluster – overlapping with sample locations that belonged to separate clusters in Lombaert et al. (2014). This may be due to the larger number of populations (16) sampled from Europe in the 2014 study compared to the four included here. It should be noted that the European and Asian native samples included in our study were collected more recently (2016–2020) than those from the 2014 study (1998–2009); our results could reflect more recent genetic mixing. Genetic structure has the potential to change over time as populations are influenced by changes in population density, gene flow and/or secondary contact, as well as additional introductions and/or admixture events (Troupin et al. 2006).
Colonisation history
Approximate Bayesian Computation (ABC) analyses suggest the most likely colonisation history of H. axyridis in our study matches the Lombaert et al. (2014) proposal that eastern North America has been a disproportionate proximate source of invasive populations of H. axyridis, sometimes referred to as a bridgehead population. Both their study and ours showed high levels of support for model scenarios where eastern North America has contributed colonists to South Africa, South America, and Greece. Rapid spread among parts of the invasive range can result in admixture among previously isolated lineages, as can pre-invasion admixture (Rius and Darling 2014). Both were clearly evident in our study, both between Asia 1 and 2 populations and South Africa between populations with proximate origins in eastern North America and an unsampled population. The finding of admixture in South Africa differs slightly from what was previously suggested by Lombaert et al. (2010; 2011; 2014). Lombaert et al. 2014’s analyses of South Africa’s invasion history suggest beetles found within South Africa are directly descendent from beetles originating from a population in eastern North America. However, we show that South Africa’s population is descendent from an admixture event between a population in eastern North America and a population not sampled in this study. However, as our North American sampling was limited, it is possible that the unsampled population predicted to have contributed individuals to South Africa’s H. axyridis population could have originated from a region in North America not sampled in this study. Lombaert et al. (2014) revealed multiple instances of genetic admixture between H. axyridis populations within North America (namely, between two populations originating from the native region, and between populations from west and east North America). Thus, genetic admixture between the two populations indicated in our analyses is likely to have occurred pre-invasion within North America. Alternatively, these beetles could have arrived as secondary invasion or could be remnants of a population of beetles that were introduced in the 1980s as biological control in the Mpumalanga Province (Roy et al. 2016). Records from this introduction suggest that the attempt to introduce the beetle had failed, however it is possible that beetles may have established but remained undetected (Roy et al. 2016). This scenario may be less likely because we found no clear evidence of multiple introductions of H. axyridis from multiple source populations in our genetic structure analyses. However, without further sampling we cannot discern among these alternative possibilities, though the accidental introduction of H. axyridis is certainly plausible as this and other coccinellid beetles have previously been recorded (often in large numbers) as stowaways on cargo and cruise ships (Sæthre et al. 2010; Brown et al. 2011; Stals 2010). One of the samples for our study included beetles intercepted at St. Helena Bay after arriving by boat from Cape Town, South Africa. These samples support the suggestion that H. axyridis beetles are making use of human-assisted transport to disperse over long distances.
The newly identified invasive population of H. axyridis in northern Greece also shows evidence of genetic admixture between two lineages, in this case eastern North America and Europe. Beetles originating from eastern North America most likely followed a similar introduction route into Greece as was described above for its introduction into South Africa, while samples from Europe most likely expanded its range from regions further North where it has established since 2001 (Brown et al. 2011). It is clear that the eastern North American outbreak of invasive beetles has played (and is most likely still playing) a critical role in the worldwide invasion of H. axyridis and is acting as a bridgehead population. Finally, populations in west North America were derived from beetles originating from eastern and western Asia, which subsequently invaded parts of Europe. Admixture was identified in European populations between beetles from western North America and an unsampled population. The unsampled population not included in our study could plausibly be the biocontrol strain that was introduced into Europe, as was found by Lombaert et al. (2014), whose study included a more extensive sampling of the European H. axyridis population. Lombaert et al. (2014) discovered multiple instances of genetic admixture within Europe, including genetic admixture between populations from western North America and a European biological control strain of H. axyridis: a strain of H. axyridis, originally collected from a native region in Asia in 1982, which was subsequently introduced into Europe and South America as a biological control agent during the 1980s (Lombaert et al. 2010; 2014).
While there were some notable changes in population structure, specifically in the native region in Asia and Europe, the temporal invasion pattern of H. axyridis has remained relatively stable since the first reports investigating its colonisation history more than ten years ago. One of the main drawbacks from our study was the limited number of sample locations procured from eastern North America to include in the population genetic structure analyses. Future studies investigating how H. axyridis in South Africa fits into the global invasion network would benefit from increasing the sampling effort in eastern North America since beetles from South Africa were probably derived from this region. However, our study was still able to reach a similar outcome as studies that included a more comprehensive coverage of samples from North America.
Genetic diversity
The populations of H. axyridis in South Africa, along with the other introduced populations included in our study (Americas and Europe), had high genetic diversity (measured as high expected heterozygosity, numbers of alleles and allelic richness) compared to populations from the native region. There are several mechanisms that may have promoted high genetic diversity within the country, namely: multiple introductions, high propagule pressure, and the genetic makeup of the source population in part derived from admixture, and de novo mutation and recombination in the context of rapid population growth. Here, we identified that South Africa’s colonisation history involved genetic admixture, which may have influenced the high genetic diversity estimates within the country. Genetic admixture has the potential to significantly influence the genetic diversity of populations by producing recombinant genotypes and creating novel sequences that may not be found in the genome of individuals within the source population (Keller and Taylor 2008), which could influence the adaptive ability of individuals in the introduced region. Furthermore, multiple introductions of non-indigenous species allow a larger subset of the genetic information within the source population to enter the new environment after each new introduction. Similarly, high propagule pressure (number of individuals introduced at single time) introduces more genetic material into the new environment during each introduction event. Multiple introductions, high propagule pressure, or a combination of both, gives rise to genetically diverse populations which assists newly colonising invasive species in counteracting genetic bottlenecks and other founder effects (Novo et al. 2015; Garnas et al. 2016; van Boheemen et al. 2017). Even though it is not clear from our genetic analyses that multiple introductions occurred within South Africa, multiple introductions from a source population are a common feature of biological invasions (Wilson et al. 2009), and previous observations of H. axyridis suggest its association with human-mediated transport channels (Brown et al. 2011), may have supported multiple introductions and/or high propagule pressure of beetles into South Africa and other regions where it was introduced. It is unclear from the genetic analyses in our study whether the introduction(s) of H. axyridis into South Africa is historical or is still ongoing. If beetles were introduced via ships, we would expect some genetic differentiation between populations closest to South Africa’s major seaport harbours (Cape Town, Port Elizabeth, East London, Durban) than those further inland. However, this is not the case. Mantel’s tests suggest that there are no significant genetic differences between populations closest to seaport harbours than those further inland. While it is still likely that beetles are currently entering the country via the sea, propagule sizes may not be large enough to affect microsatellite genetic diversity in the regions surrounding seaport harbours.
Recent bottlenecks were detected in 12 out of 14 South African populations. This may seem contradictory to the high genetic diversity estimates found for all South African populations, as genetic bottlenecks are often associated with decreased genetic diversity (Garnas et al. 2016). However, it is not uncommon for introduced populations to experience short-lived bottlenecks lasting only a few generations, followed by rapid population expansions (Luikart and Cornuet 1998). Genetic bottlenecks can also, under certain conditions, lead to an increase in genetic variance provided that epistatic variance is converted to additive variance (Naciri-Graven and Goudet 2003). An alternative hypothesis could be that prior to the genetic bottleneck, the population had high genetic diversity to begin with. Previous studies investigating high genetic diversity succeeding genetic bottlenecks suggested this may be the case for the short-tailed albatross, Phoebastria albatrus, (Kuro-o et al. 2010), and brown seaweed, Saccorhiza polyschides (Assis et al. 2013). This may be the most likely explanation for H. axyridis beetles within South Africa owing to the genetic admixture in its invasion history and the potential effects it has had on genetic diversity (hybrid vigor, purging deleterious mutations; Facon et al. 2011; Turgeon et al. 2011; Hahn and Rieseberg 2017).
Conclusion
The spread of non-native species is most often facilitated by unintentional introductions, which frequently go undetected and as a result spread faster than those introduced intentionally (Hill et al. 2016; Roques et al. 2016). Our findings suggest that the unintentional introduction(s) of H. axyridis from eastern North America acted as the source of colonists invading South Africa, parts of South America and Europe, in conjunction with an undetected source. As suggested by Lombaert et al. (2010), bridgehead populations should receive increased vigilance to manage the spread of invasive species. Several invasive species have involved, at least in some way, secondary introductions from bridgehead populations (Bertlesmeier et al 2018; Correa et al. 2019; Javal et al. 2018). After ten years since the initial article investigating the worldwide colonisation history of H. axyridis was published, our study suggests that the invasion patterns have remained largely stable with minimal changes in population structure and patterns and rates of gene flow. This is perhaps surprising since one might expect increasing control, improved management and better prevention of introductions as social awareness of the invasive species grows. Furthermore, H. axyridis beetles appear to be to be experiencing high connectivity both globally and across South Africa. Management plans would therefore have to consider H. axyridis’ high connectivity when assessing potential control/management strategies for this species and at what scale it should be implemented, and would perhaps need a multi-faceted or integrated approach.
Data availability
The microsatellite data generated during the current study were deposited in the Zenodo open repository and is available at the following URL: https://doi.org/https://doi.org/10.5281/zenodo.7953069.
References
Assis J, Castilho Coelho N, Alberto F, Valero M, Raimondi P, Reed D, Alvares Serrão E (2013) High and distinct range-edge genetic diversity despite local bottlenecks. PLoS ONE 8(7):e68646
Belkhir K, Borsa P, Chikhi L, Raufaste N, Bonhomme F (2004) GENETIX 4.05, Population genetics software for Windows TM. Université de Montpellier II. Montpellier.
Bertelsmeier C, Ollier S, Liebhold AM, Brockerhoff EG, Ward D, Keller L (2018) Recurrent bridgehead effects accelerate global alien ant spread. Proc Natl Acad Sci 115(21):5486–5491
Bock DG, Caseys C, Cousens RD, Hahn MA, Heredia SM, Hübner S, Turner KG, Whitney KD, Rieseberg LH (2015) What we still don’t know about invasion genetics. Mol Ecol 24(9):2277–2297
Bowcock AM, Ruiz-Linares A, Tomfohrde J, Minch E, Kidd JR, Cavalli-Sforza LL (1994) High resolution of human evolutionary trees with polymorphic microsatellites. Nature 368(6470):455–457
Brown PM, Thomas CE, Lombaert E, Jeffries DL, Estoup A, Lawson Handley LJ (2011) The global spread of Harmonia axyridis (Coleoptera: Coccinellidae): distribution, dispersal and routes of invasion. Biocontrol 56(4):623–641
Caley P, Ingram R, De Barro P (2015) Entry of exotic insects into Australia: does border interception count match incursion risk? Biol Invasions 17(4):1087–1094
Camacho-Cervantes M, Ortega-Iturriaga A, Del-Val E (2017) From effective biocontrol agent to successful invader: the harlequin ladybird (Harmonia axyridis) as an example of good ideas that could go wrong. PeerJ 5:e3296
Ceryngier P, Romanowski J (2017) Harmonia axyridis (Pallas, 1773)(Coleoptera: Coccinellidae) and its parasite in south-western Bulgaria and northern Greece. BioInvasions Records 6:307–310
Chapin JB, Brou VA (1991) Harmonia axyridis (Pallas), the third species of the genus to be found in the United States (Coleoptera: Coccinellidae). Proc Entomol Soc Wash 93(3):630–635
Chapuis MP, Estoup A (2007) Microsatellite null alleles and estimation of population differentiation. Mol Biol Evol 24(3):621–631
Ciosi M, Miller NJ, Toepfer S, Estoup A, Guillemaud T (2011) Stratified dispersal and increasing genetic variation during the invasion of Central Europe by the western corn rootworm. Diabrotica Virgifera Virgifera Evolutionary Appl 4(1):54–70
Cornuet JM, Luikart G (1996) Description and power analysis of two tests for detecting recent population bottlenecks from allele frequency data. Genetics 144(4):2001–2014
Cornuet JM, Pudlo P, Veyssier J, Dehne-Garcia A, Gautier M, Leblois R, Marin JM, Estoup A (2014) DIYABC v2. 0: a software to make approximate Bayesian computation inferences about population history using single nucleotide polymorphism DNA sequence and microsatellite data. Bioinformatics 30(8):1187–1189
Correa MC, Palero F, Malausa T, Crochard D, Zaviezo T, Lombaert E (2019) European bridgehead effect in the worldwide invasion of the obscure mealybug. Biol Invasions 21(1):123–136
Estoup A, Guillemaud T (2010) Reconstructing routes of invasion using genetic data: why, how and so what? Mol Ecol 19(19):4113–4130
Evanno G, Regnaut S, Goudet J (2005) Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol 14(8):2611–2620
Excoffier L, Lischer HE (2010) Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Mol Ecol Resour 10(3):564–567
Facon B, Hufbauer RA, Tayeh A, Loiseau A, Lombaert E, Vitalis R, Guillemaud T, Lundgren JG, Estoup A (2011) Inbreeding depression is purged in the invasive insect Harmonia axyridis. Curr Biol 21(5):424–427
Falush D, Stephens M, Pritchard JK (2003) Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics 164(4):1567–1587
Galvan TL, Koch RL, Hutchison WD (2008) Impact of fruit feeding on overwintering survival of the multicolored Asian lady beetle, and the ability of this insect and paper wasps to injure wine grape berries. Entomol Exp Appl 128(3):429–436
Garnas JR (2018) Rapid evolution of insects to global environmental change: conceptual issues and empirical gaps. Curr Opin Insect Sci 29:93–101
Garnas J, Auger-Rozenberg MA, Roques A, Bertelsmeier C, Wingfield MJ, Saccaggi DL, Roy HE, Slippers B (2016) Complex patterns of global spread in invasive insects: eco-evolutionary and management consequences. Biol Invasions 18(4):935–952
Gilbert M, Grégoire JC, Freise JF, Heitland W (2004) Long-distance dispersal and human population density allow the prediction of invasive patterns in the horse chestnut leafminer Cameraria ohridella. J Anim Ecol 73(3):459–468
Goudet J (2001) FSTAT, a program to estimate and test gene diversities and fixation indices, version 2.9.3. http://www2.unil.ch/popgen/softwares/fstat.htm
Goudet J, Jombart T (2015) hierfstat: estimation and tests of hierarchical F-statistics. R package version 0.04–22, 10.
Hahn MA, Rieseberg LH (2017) Genetic admixture and heterosis may enhance the invasiveness of common ragweed. Evol Appl 10(3):241–250
Hill MP, Clusella-Trullas S, Terblanche JS, Richardson DM (2016) Drivers, impacts, mechanisms and adaptation in insect invasions. Biol Invasions 18(4):883–891
Hufbauer RA, Facon B, Ravigné V, Turgeon J, Foucaud J, Lee CE, Rey O, Estoup A (2012) Anthropogenically induced adaptation to invade (AIAI): contemporary adaptation to human-altered habitats within the native range can promote invasions. Evol Appl 5(1):89–101
Iablokoff-Khnzorian S M (1982) Les coccinelles, Coléopteres-Coccinellidae: tribu coccinellini des régions paléarctique et orientale. Éditions Boubée.
Javal M, Roux G, Roques A, Sauvard D (2018) Asian Long-horned Beetle dispersal potential estimated in computer-linked flight mills. J Appl Entomol 142(1–2):282–286
Jeffries DL, Chapman J, Roy HE, Humphries S, Harrington R, Brown PM, Handley LJL (2013) Characteristics and drivers of high-altitude ladybird flight: insights from vertical-looking entomological radar. PLoS ONE 8(12):e82278
Jombart T, Ahmed I (2011) Adegenet 1.3–1: new tools for the analysis of genome-wide SNP data. Bioinformatics 27(21):3070–3071
Karsten M, Jansen van Vuuren B, Addison P, Terblanche JS (2015) Deconstructing intercontinental invasion pathway hypotheses of the Mediterranean fruit fly (Ceratitis capitata) using a Bayesian inference approach: are port interceptions and quarantine protocols successfully preventing new invasions? Divers Distrib 21(7):813–825
Karsten M, Barton M, Addison P, Addison MF, Terblanche JS (2018) A synthesis for managing invasions and pest risks simultaneously for tephritid fruit flies in South Africa. Entomol Exp Appl 166(5):344–356
Keller R, Taylor DR (2008) History, chance and adaptation during biological invasion: separating stochastic phenotypic evolution from response to selection. Ecol Lett 11(8):852–866
Kenis M, Rabitsch W, Auger-Rozenberg MA, Roques A (2007) How can alien species inventories and interception data help us prevent insect invasions? Bull Entomol Res 97(5):489–502
Koch RL (2003) The multicolored Asian lady beetle, Harmonia axyridis: a review of its biology, uses in biological control, and non-target impacts. J Insect Sci 3(1):32
Kopelman N M, Mayzel J, Jakobsson M, Rosenberg N A, Mayrose I (2015) Clumpak: a program for identifying clustering modes and packaging population structure inferences across K. Mol Ecol Resour 15(5):1179–1191
Kuro-o M, Yonekawa H, Saito S, Eda M, Higuchi H, Koike H, Hasegawa H (2010) Unexpectedly high genetic diversity of mtDNA control region through severe bottleneck in vulnerable albatross Phoebastria albatrus. Conserv Genet 11(1):127–137
Lockwood JL, Cassey P, Blackburn T (2005) The role of propagule pressure in explaining species invasions. Trends Ecol Evol 20(5):223–228
Loiseau A, Malausa T, Lombaert E, MARTIN J F, Estoup A (2009) Isolation and characterization of microsatellites in the harlequin ladybird, Harmonia axyridis (Coleoptera, Coccinellidae), and cross‐species amplification within the family Coccinellidae. Mol Ecol Resour 9(3):934–937
Lombaert E, Guillemaud T, Cornuet JM, Malausa T, Facon B, Estoup A (2010) Bridgehead effect in the worldwide invasion of the biocontrol harlequin ladybird. PLoS ONE 5(3):e9743
Lombaert E, Guillemaud T, Thomas CE, Lawson Handley LJ, Li J, Wang S, Pang H, Goryacheva I, Zakharov IA, Jousselin E, Poland RL (2011) Inferring the origin of populations introduced from a genetically structured native range by approximate Bayesian computation: case study of the invasive ladybird Harmonia axyridis. Mol Ecol 20(22):4654–4670
Lombaert E, Guillemaud T, Lundgren J, Koch R, Facon B, Grez A, Loomans A, Malausa T, Nedved O, Rhule E, Staverlokk A (2014) Complementarity of statistical treatments to reconstruct worldwide routes of invasion: the case of the Asian ladybird Harmonia axyridis. Mol Ecol 23(24):5979–5997
Luikart G, Cornuet JM (1998) Empirical evaluation of a test for identifying recently bottlenecked populations from allele frequency data. Conserv Biol 12(1):228–237
Michaud JP (2002) Invasion of the Florida citrus ecosystem by Harmonia axyridis (Coleoptera: Coccinellidae) and asymmetric competition with a native species. Cycloneda Sanguinea Environ Entomol 31(5):827–835
Naciri-Graven Y, Goudet J (2003) The additive genetic variance after bottlenecks is affected by the number of loci involved in epistatic interactions. Evolution 57(4):706–716
Nedvěd O, Háva J (2016) New record of the invasive ladybeetle Harmonia axyridis in Afrotropical Region: Tanzania. Zanzibar African Entomol 24(1):247–249
Novo M, Cunha L, Maceda-Veiga A, Talavera JA, Hodson ME, Spurgeon D, Bruford MW, Morgan AJ, Kille P (2015) Multiple introductions and environmental factors affecting the establishment of invasive species on a volcanic island. Soil Biol Biochem 85:89–100
Osawa N (2000) Population field studies on the aphidophagous ladybird beetle Harmonia axyridis (Coleoptera: Coccinellidae): resource tracking and population characteristics. Popul Ecol 42(2):115–127
Peakall R, Smouse PE (2006) GENALEX 6: genetic analysis in Excel. Population genetic software for teaching and research. Mol Ecol Notes 6:288–295
Peakall R, Smouse PE (2012) GENALEX 6.5: genetic analysis in Excel. Population genetic software for teaching and research – an update. Bioinformatics 28:2537–2539
Pickering G, Lin J, Riesen R, Reynolds A, Brindle I, Soleas G (2004) Influence of Harmonia axyridis on the sensory properties of white and red wine. Am J Enol Vitic 55(2):153–159
Poutsma J, Loomans A J M, Aukema B, Heijerman T (2007) Predicting the potential geographical distribution of the harlequin ladybird, Harmonia axyridis, using the CLIMEX model. In: From Biological Control to Invasion: the ladybird Harmonia axyridis as a model species. Springer, Dordrecht, pp. 103–125)
Pratt PD, Center TD (2012) Biocontrol without borders: the unintended spread of introduced weed biological control agents. Biocontrol 57(2):319–329
Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155(2):945–959
Raak-van den Berg CL, Hemerik L, de Jong PW, van Lenteren JC (2012) Mode of overwintering of invasive Harmonia axyridis in the Netherlands. Biocontrol 57(1):71–84
Ramakrishnan UMA, Hadly EA, Mountain JL (2005) Detecting past population bottlenecks using temporal genetic data. Mol Ecol 14(10):2915–2922
Rambaut A (2010) FigTree v1.3.1. Institute of Evolutionary Biology, University of Edinburgh, Edinburgh. http://tree.bio.ed.ac.uk/software/figtree/.
Rius M, Darling JA (2014) How important is intraspecific genetic admixture to the success of colonising populations? Trends Ecol Evol 29(4):233–242
Roques A, Auger-Rozenberg MA, Blackburn TM, Garnas J, Pyšek P, Rabitsch W, Richardson DM, Wingfield MJ, Liebhold AM, Duncan RP (2016) Temporal and interspecific variation in rates of spread for insect species invading Europe during the last 200 years. Biol Invasions 18(4):907–920
Rousset F (2008) genepop’007: a complete re‐implementation of the genepop software for Windows and Linux. Mol Ecol Resour 8(1):103–106
Roy HE, Brown PM (2015) Ten years of invasion: Harmonia axyridis (Pallas)(Coleoptera: Coccinellidae) in Britain. Ecological Entomology 40(4):336–348
Roy HE, Brown PM, Adriaens T, Berkvens N, Borges I, Clusella-Trullas S, Comont RF, De Clercq P, Eschen R, Estoup A, Evans EW et al (2016) The harlequin ladybird, Harmonia axyridis: global perspectives on invasion history and ecology. Biol Invasions 18(4):997–1044
Sæthre MG, Staverløkk A, Hofsvang T (2010) The history of Harmonia axyridis Pallas 1773 in Norway. Iobc/wprs Bull 58:97–104
Seebens H, Blackburn TM, Dyer EE, Genovesi P, Hulme PE, Jeschke JM, Pagad S, Pyšek P, Winter M, Arianoutsou M, Bacher S et al (2017) No saturation in the accumulation of alien species worldwide. Nat Commun 8(1):1–9
Seko T, Yamashita KI, Miura K (2008) Residence period of a flightless strain of the ladybird beetle Harmonia axyridis Pallas (Coleoptera: Coccinellidae) in open fields. Biol Control 47(2):194–198
Stals R (2010) The establishment and rapid spread of an alien invasive lady beetle: Harmonia axyridis (Coleoptera: Coccinellidae) in southern Africa, 2001–2009. Benefits Risks Exotic Biol Agents 58:125–132
Stals R, Prinsloo G (2007) Discovery of an alien invasive predatory insect in South Africa: the multi-coloured Asian ladybird beetle, Harmonia axyridis (Pallas) (Coleoptera: Coccinellidae). Int Organ Biol Integrated Control/ West Palearctic Regional Sect Bulletin 103:123–126
Takezaki N, Nei M, Tamura K (2014) Poptreew: web version of Poptree for constructing population trees from allele frequency data and computing some other quantities. Mol Biol Evol 31:1625–1637
Troupin, D., Nathan, R., & Vendramin, G. G. (2006). Analysis of spatial genetic structure in an expanding Pinus halepensis population reveals development of fine‐scale genetic clustering over time. Mol Ecol 15(12):3617–3630
Turgeon J, Tayeh A, Facon B, Lombaert E, De Clercq P, Berkvens N, Lundgren G, Estoup A (2011) Experimental evidence for the phenotypic impact of admixture between wild and biocontrol Asian ladybird (Harmonia axyridis) involved in the European invasion. J Evol Biol 24(5):1044–1052
van Boheemen LA, Lombaert E, Nurkowski KA, Gauffre B, Rieseberg LH, Hodgins KA (2017) Multiple introductions, admixture and bridgehead invasion characterize the introduction history of Ambrosia artemisiifolia in Europe and Australia. Mol Ecol 26(20):5421–5434
Van Wilgen BW, Measey J, Richardson DM, Wilson JR, Zengeya TA (2020) Biological Invasions in South Africa. Springer, Springer Nature
Weir BS, Cockerham CC (1984) Estimating F-statistics for the analysis of population structure. Evolution 38:1358–1370
Wilson JRU, Domontt EE, Prentis PJ, Lowe AJ, Richardson DM (2009) Something in the way you move: dispersal pathways affect invasion success. Trends Ecol 24:136–144
Wright S (1946) Isolation by distance under diverse systems of mating. Genetics 31(1):39
Acknowledgements
We thank Dmitry Musolin, Riaan Stals, Qi Xuewei, Niu Changying, James Menger-Anderson, Elizabeth Gazzano, Spencer Behmer, Marc De Meyer and Ingrid Minnaar for contributing to the Harlequin Ladybird citizen science project or providing additional samples. We are grateful to Marion Javal and Anandi Bierman for technical support and discussion. We thank referees and examiners for their valuable comments and suggestions on earlier versions of this work. The microsatellite data were genotyped at the Central Analytical Facility (CAF). This work was funded by the DSI-NRF Centre of Excellence for Invasion Biology (CIB), Stellenbosch University, South Africa, to Amy Collop, Susana Clusella-Trullas and John S. Terblanche.
Funding
Open access funding provided by Stellenbosch University. This work was funded by the DSI-NRF Centre of Excellence for Invasion Biology (CIB) to Amy Collop, Susana Clusella-Trullas and John S. Terblanche.
Author information
Authors and Affiliations
Contributions
Minette Karsten, John S. Terblanche, Susana Clusella-Trullas contributed to the study conception and design. Material preparation, data collection and analysis were performed by Amy Collop and Minette Karsten. The first draft of the manuscript was written by Amy Collop and all authors commented on subsequent versions of the manuscript. All authors read and approved the final version of the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors have no relevant financial or non-financial interests to disclose.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Collop, A., Terblanche, J.S., Clusella-Trullas, S. et al. Testing hypotheses of invasion pathways of the ladybird Harmonia axyridis into and across South Africa. Biol Invasions 26, 1715–1734 (2024). https://doi.org/10.1007/s10530-024-03274-w
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10530-024-03274-w