Abstract
Purpose
Purple acid phosphatases (PAPs) includ the largest classes of non-specific plant acid phosphatases. Most characterized PAPs were found to play physiological functions in phosphorus metabolism. In this study, we investigated the function of AtPAP17 gene encoding an important purple acid phosphatase in Arabidopsis thaliana.
Methods
The full-length cDNA sequence of AtPAP17 gene under the control of CaMV-35S promoter was transferred to the A. thaliana WT plant. The generated homozygote AtPAP17-overexpressed plants were compared by the types of analyses with corresponding homozygote atpap17-mutant plant and WT in both + P (1.2 mM) and − P (0 mM) conditions.
Results
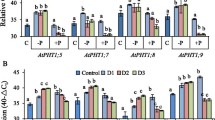
In the + P condition, the highest and the lowest amount of Pi was observed in AtPAP17-overexpressed plants and atpap17-mutant plants by 111% increase and 38% decrease compared with the WT plants, respectively. Furthermore, under the same condition, APase activity of AtPAP17-overexpressed plants increased by 24% compared to the WT. Inversely, atpap17-mutant plant represented a 71% fall compared to WT plants. The comparison of fresh weight and dry weight in the studied plants showed that the highest and the lowest amount of absorbed water belonged to OE plants (with 38 and 12 mg plant−1) and Mu plants (with 22 and 7 mg plant−1) in + P and − P conditions, respectively.
Conclusion
The lack of AtPAP17 gene in the A. thaliana genome led to a remarkable reduction in the development of root biomass. Thus, AtPAP17 could have an important role in the root but not shoot developmental and structural programming. Consequently, this function enables them to absorb more water and eventually associated with more phosphate absorption.






Similar content being viewed by others
Data availability
All data generated or analyzed during this study are included in the article/supplementary material, further inquiries can be directed to the corresponding author/s.
References
Abbasi-Vineh MA, Sabet MS, Karimzadeh G (2021) Identification and functional analysis of two purple acid phosphatases AtPAP17 and AtPAP26 involved in salt tolerance in Arabidopsis thaliana plant. Front Plant Sci 11:618716. https://doi.org/10.3389/fpls.2020.618716
Ames BN (1966) Assay of inorganic phosphate, total phosphate and phosphatases. In: Neufeld EF, Ginsburg V (eds) Methods in enzymology, vol 8. Academic Press, New York, pp 115–118. https://doi.org/10.1016/0076-6879(66)08014-5
Bechtold N, Pelletier G (1998) In planta Agrobacterium mediated transformation of adult Arabidopsis thaliana plants by vacuum Infiltration. In: Martinez-Zapater JM, Salinas J (eds) Arabidopsis protocols. Humana Press, Totowa, pp 259–266
Benkert P, Tosatto SCE, Schombu D (2008) QMEAN: a comprehensive scoring function for model quality assessment. Proteins 71:261–277. https://doi.org/10.1002/prot.21715
Berman HM, Westbrook J, Feng Z et al (2000) The protein data bank. Nucleic Acids Res 28:235–242. https://doi.org/10.1093/nar/28.1.235
Biasini M, Bienert S, Waterhouse A et al (2014) SWISS-MODEL: modelling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Res 42:W252–W258. https://doi.org/10.1093/nar/gku340
Birnboim HC, Doly J (1979) A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res 7:1513–1523. https://doi.org/10.1093/nar/7.6.1513
Bowie JU, Lüthy R, Eisenberg D (1991) A method to identify protein sequences that fold into a known three-dimensional structure. Science 253:164–170. https://doi.org/10.1126/science.1853201
Bradford MM (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72:248–254. https://doi.org/10.1016/0003-2697(76)90527-3
Burley SK, Berman HM, Bhikadiya C et al (2019) RCSB protein data bank: biological macromolecular structures enabling research and education in fundamental biology, biomedicine, biotechnology and energy. Nucleic Acids Res 47:D464–D474. https://doi.org/10.1093/nar/gky1004
Bustos R, Castrillo G, Linhares F et al (2010) A central regulatory system largely controls transcriptional activation and repression responses to phosphate starvation in Arabidopsis. PLoS Genet 6:e1001102. https://doi.org/10.1371/journal.pgen.1001102
Camacho C, Coulouris G, Avagyan V et al (2009) BLAST+: architecture and applications. BMC Bioinform 10:421. https://doi.org/10.1186/1471-2105-10-421
Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16:735–743. https://doi.org/10.1046/j.1365-313x.1998.00343.x
Colovos C, Yeates TO (1993) Verification of protein structures: patterns of nonbonded atomic interactions. Protein Sci 2:1511–1519. https://doi.org/10.1002/pro.5560020916
Coutsias EA, Seok C, Jacobson MP, Dill KA (2004) A kinematic view of loop closure. J Comput Chem 25:510–528. https://doi.org/10.1002/jcc.10416
Czarnecki O, Yang J, Weston DJ et al (2013) A dual role of strigolactones in phosphate acquisition and utilization in plants. Int J Mol Sci 14:7681–7701. https://doi.org/10.3390/ijms14047681
Del Pozo JC, Allona I, Rubio V et al (1999) A type 5 acid phosphatase gene from Arabidopsis thaliana is induced by phosphate starvation and by some other types of phosphate mobilizing/oxidative stress conditions. Plant J 19:579–589. https://doi.org/10.1046/j.1365-313X.1999.00562.x
Del Vecchio HA, Ying S, Park J et al (2014) The cell wall-targeted purple acid phosphatase AtPAP25 is critical for acclimation of Arabidopsis thaliana to nutritional phosphorus deprivation. Plant J 80:569–581. https://doi.org/10.1111/tpj.12663
Delano W (2015) The PyMOL molecular graphics system. Schrodinger, V 2.0.7. http://www.pymol.sourceforge.net
Farhadi S, Sabet MS, Malboobi MA, Moieni A (2020) The critical role of AtPAP17 and AtPAP26 genes in Arabidopsis phosphate compensation network. Front Plant Sci 11:565865. https://doi.org/10.3389/fpls.2020.565865
Feder D, McGeary RP, Mitić N et al (2020) Structural elements that modulate the substrate specificity of plant purple acid phosphatases: avenues for improved phosphorus acquisition in crops. Plant Sci 294:110445. https://doi.org/10.1016/j.plantsci.2020.110445
Fernandez-Fuentes N, Madrid-Aliste CJ, Rai BK et al (2007) M4T: a comparative protein structure modeling server. Nucleic Acids Res 35:W363–W368. https://doi.org/10.1093/nar/gkm341
Finn RD, Coggill P, Eberhardt RY et al (2016) The Pfam protein families database: towards a more sustainable future. Nucleic Acids Res 44:D279–D285. https://doi.org/10.1093/nar/gkv1344
Flanagan JU, Cassady AI, Schenk G et al (2006) Identification and molecular modeling of a novel, plant-like, human purple acid phosphatase. Gene 377:12–20. https://doi.org/10.1016/j.gene.2006.02.031
Gahan LR, Smith SJ, Neves A, Schenk G (2009) Phosphate ester hydrolysis: metal complexes as purple acid phosphatase and phosphotriesterase analogues. Eur J Inorg Chem 2009:2745–2758. https://doi.org/10.1002/ejic.200900231
Ghahremani M, Tran H, Biglou SG et al (2019) A glycoform of the secreted purple acid phosphatase AtPAP26 co-purifies with a mannose-binding lectin (AtGAL1) up-regulated by phosphate-starved Arabidopsis. Plant Cell Environ 42:1139–1157. https://doi.org/10.1111/pce.13432
Greenway H, Klepper B (1968) Phosphorus transport to the xylem and its regulation by water flow. Planta 83:119–136. https://doi.org/10.1007/BF00385017
Guex N, Peitsch MC (1997) SWISS-MODEL and the Swiss-Pdb Viewer: an environment for comparative protein modeling. Electrophoresis 18:2714–2723. https://doi.org/10.1002/elps.1150181505
Heinonen JK, Lahti RJ (1981) A new and convenient colorimetric determination of inorganic orthophosphate and its application to the assay of inorganic pyrophosphatase. Anal Biochem 113:313–317. https://doi.org/10.1016/0003-2697(81)90082-8
Heo L, Park H, Seok C (2013) GalaxyRefine: protein structure refinement driven by side-chain repacking. Nucleic Acids Res 41:W384–W388. https://doi.org/10.1093/nar/gkt458
Hoagland DR, Arnon DI (1950) The water-culture method for growing plants without soil, 2nd edn. California Agricultural Experiment Station, Berkeley, California, pp 1–32
Holsters M, de Waele D, Depicker A et al (1978) Transfection and transformation of Agrobacterium tumefaciens. Mol Gen Genet 163:181–187. https://doi.org/10.1007/BF00267408
Hurley BA, Tran HT, Marty NJ et al (2010) The dual-targeted purple acid phosphatase isozyme AtPAP26 is essential for efficient acclimation of Arabidopsis to nutritional phosphate deprivation. Plant Physiol 153:1112–1122. https://doi.org/10.1104/pp.110.153270
Kaida R, Hayashi T, Kaneko TS (2008) Purple acid phosphatase in the walls of tobacco cells. Phytochemistry 69:2546–2551. https://doi.org/10.1016/j.phytochem.2008.07.008
Källberg M, Wang H, Wang S et al (2012) Template-based protein structure modeling using the RaptorX web server. Nat Protoc 7:1511–1522. https://doi.org/10.1038/nprot.2012.085
Kelley LA, Mezulis S, Yates CM et al (2015) The Phyre2 web portal for protein modeling, prediction and analysis. Nat Protoc 10:845–858. https://doi.org/10.1038/nprot.2015.053
Ko J, Park H, Heo L, Seok C (2012) GalaxyWEB server for protein structure prediction and refinement. Nucleic Acids Res 40:W294–W297. https://doi.org/10.1093/nar/gks493
Krysan PJ, Young JC, Sussman MR (1999) T-DNA as an insertional mutagen in Arabidopsis. Plant Cell 11:2283–2290. https://doi.org/10.1105/tpc.11.12.2283
Kuang R, Chan KH, Yeung E, Lim BL (2009) Molecular and biochemical characterization of AtPAP15, a purple acid phosphatase with phytase activity, in Arabidopsis. Plant Physiol 151:199–209. https://doi.org/10.1104/pp.109.143180
Kumar K, Yadava P, Gupta M et al (2022) Narrowing down molecular targets for improving phosphorus-use efficiency in maize (Zea mays L.). Mol Biol Rep 49:12091–12107. https://doi.org/10.1007/s11033-022-07679-5
Laskowski RA, MacArthur MW, Moss DS, Thornton JM (1993) PROCHECK: a program to check the stereochemical quality of protein structures. J Appl Crystallogr 26:283–291. https://doi.org/10.1107/S0021889892009944
Laskowski RA, Rullmann JAC, MacArthur MW et al (1996) AQUA and PROCHECK-NMR: programs for checking the quality of protein structures solved by NMR. J Biomol NMR 8:477–486. https://doi.org/10.1007/BF00228148
Lee GR, Heo L, Seok C (2016) Effective protein model structure refinement by loop modeling and overall relaxation. Proteins 84:293–301. https://doi.org/10.1002/prot.24858
Li D, Zhu H, Liu K et al (2002) Purple acid phosphatases of Arabidopsis thaliana. Comparative analysis and differential regulation by phosphate deprivation. J Biol Chem 277:27772–27781. https://doi.org/10.1074/jbc.M204183200
Lin W-Y, Lin S-I, Chiou T-J (2009) Molecular regulators of phosphate homeostasis in plants. J Exp Bot 60:1427–1438. https://doi.org/10.1093/jxb/ern303
Lovell SC, Davis IW, Arendall WB III et al (2003) Structure validation by Cα geometry: ϕ, ψ and Cβ deviation. Proteins 50:437–450. https://doi.org/10.1002/prot.10286
Lu K, Li J, Zhong W et al (2008) Isolation, characterization and phosphate-starvation inducible expression of potential Brassica napus purple acid phosphatase 17 (BnPAP17) gene family. Bot Stud 17:199–213
Lüthy R, Bowie JU, Eisenberg D (1992) Assessment of protein models with three-dimensional profiles. Nature 356:83–85. https://doi.org/10.1038/356083a0
Ma J, Peng J, Wang S, Xu J (2012) A conditional neural fields model for protein threading. Bioinformatics 28:i59–i66. https://doi.org/10.1093/bioinformatics/bts213
McGuffin LJ, Atkins JD, Salehe BR et al (2015) IntFOLD: an integrated server for modelling protein structures and functions from amino acid sequences. Nucleic Acids Res 43:W169–W173. https://doi.org/10.1093/nar/gkv236
Mehta D, Ghahremani M, Pérez-Fernández M et al (2021) Phosphate and phosphite have a differential impact on the proteome and phosphoproteome of Arabidopsis suspension cell cultures. Plant J 105:924–941. https://doi.org/10.1111/tpj.15078
Mukatira UT, Liu C, Varadarajan DK, Raghothama KG (2001) Negative regulation of phosphate starvation-induced genes. Plant Physiol 127:1854–1862. https://doi.org/10.1104/pp.010876
Murashige T, Skoog F (1962) A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiol Plant 15:473–497. https://doi.org/10.1111/j.1399-3054.1962.tb08052.x
Naseri JI, Truong NT, Hörentrup J et al (2004) Porcine purple acid phosphatase: heterologous expression, characterization, and proteolytic analysis. Arch Biochem Biophys 432:25–36. https://doi.org/10.1016/j.abb.2004.08.008
O’Gallagher B, Ghahremani M, Stigter K et al (2022) Arabidopsis PAP17 is a dual-localized purple acid phosphatase up-regulated during phosphate deprivation, senescence, and oxidative stress. J Exp Bot 73:382–399. https://doi.org/10.1093/jxb/erab409
O’Malley RC, Barragan CC, Ecker JR (2015) A user’s guide to the Arabidopsis T-DNA insertion mutant collections. In: Alonso JM, Stepanova AN (eds) Plant functional genomics: methods and protocols. Springer, New York, pp 323–342
Parinov S, Sundaresan V (2000) Functional genomics in Arabidopsis: large-scale insertional mutagenesis complements the genome sequencing project. Curr Opin Biotechnol 11:157–161. https://doi.org/10.1016/S0958-1669(00)00075-6
Park H, Seok C (2012) Refinement of unreliable local regions in template-based protein models. Proteins 80:1974–1986. https://doi.org/10.1002/prot.24086
Park H, Ko J, Joo K et al (2011) Refinement of protein termini in template-based modeling using conformational space annealing. Proteins 79:2725–2734. https://doi.org/10.1002/prot.23101
Park H, Lee GR, Heo L, Seok C (2014) Protein loop modeling using a new hybrid energy function and its application to modeling in inaccurate structural environments. PLoS ONE 9:e113811. https://doi.org/10.1371/journal.pone.0113811
Pieper U, Webb BM, Dong GQ et al (2014) ModBase, a database of annotated comparative protein structure models and associated resources. Nucleic Acids Res 42:D336–D346. https://doi.org/10.1093/nar/gkt1144
Plaxton WC (2004) Plant response to stress: biochemical adaptations to phosphate deficiency. In: Goodman RM (ed) Encyclopedia of plant and crop science. Marcel Dekker, New York, pp 976–980. https://doi.org/10.1081/E-EPCS-120010648
Pundir S, Martin MJ, O’Donovan C (2017) UniProt protein knowledgebase. In: Wu CH, Arighi CN, Ross KE (eds) Protein bioinformatics: from protein modifications and networks to proteomics. Springer, New York, pp 41–55
Radhamony RN, Mohan Prasad A, Srinivasan R (2005) T-DNA insertional mutagenesis in Arabidopsis: a tool for functional genomics. Electron J Biotechnol 8:82–106
Ravichandran S, Stone SL, Benkel B, Prithiviraj B (2013) Purple acid phosphatase 5 is required for maintaining basal resistance against Pseudomonas syringae in Arabidopsis. BMC Plant Biol 13:107. https://doi.org/10.1186/1471-2229-13-107
Remmert M, Biegert A, Hauser A, Söding J (2012) HHblits: lightning-fast iterative protein sequence searching by HMM-HMM alignment. Nat Methods 9:173–175. https://doi.org/10.1038/nmeth.1818
Rhee SY, Beavis W, Berardini TZ et al (2003) The Arabidopsis information resource (TAIR): a model organism database providing a centralized, curated gateway to Arabidopsis biology, research materials and community. Nucleic Acids Res 31:224–228. https://doi.org/10.1093/nar/gkg076
Robinson WD, Carson I, Ying S et al (2012a) Eliminating the purple acid phosphatase AtPAP26 in Arabidopsis thaliana delays leaf senescence and impairs phosphorus remobilization. New Phytol 196:1024–1029. https://doi.org/10.1111/nph.12006
Robinson WD, Park J, Tran HT et al (2012b) The secreted purple acid phosphatase isozymes AtPAP12 and AtPAP26 play a pivotal role in extracellular phosphate-scavenging by Arabidopsis thaliana. J Exp Bot 63:6531–6542. https://doi.org/10.1093/jxb/ers309
Rogers SO, Bendich AJ (1985) Extraction of DNA from milligram amounts of fresh, herbarium and mummified plant tissues. Plant Mol Biol 5:69–76. https://doi.org/10.1007/BF00020088
Sabet MS, Zamani K, Lohrasebi T et al (2018) Functional assessment of an overexpressed Arabidopsis purple acid phosphatase gene (AtPAP26) in tobacco plants. Iran J Biotechnol 16(1):31–41. https://doi.org/10.21859/ijb.2024
Schachtman DP, Reid RJ, Ayling SM (1998) Phosphorus uptake by plants: from soil to cell. Plant Physiol 116:447–453. https://doi.org/10.1104/pp.116.2.447
Schenk G, Guddat LW, Ge Y et al (2000) Identification of mammalian-like purple acid phosphatases in a wide range of plants. Gene 250:117–125. https://doi.org/10.1016/S0378-1119(00)00186-4
Schenk G, Mitić N, Hanson GR, Comba P (2013) Purple acid phosphatase: a journey into the function and mechanism of a colorful enzyme. Coord Chem Rev 257:473–482. https://doi.org/10.1016/j.ccr.2012.03.020
Schneider CA, Rasband WS, Eliceiri KW (2012) NIH image to ImageJ: 25 years of image analysis. Nat Methods 9:671–675. https://doi.org/10.1038/nmeth.2089
Sezonov G, Joseleau-Petit D, D’Ari R (2007) Escherichia coli physiology in luria-bertani broth. J Bacteriol 189:8746–8749. https://doi.org/10.1128/JB.01368-07
Srivastava R, Akash PAP et al (2020) Identification, structure analysis, and transcript profiling of purple acid phosphatases under Pi deficiency in tomato (Solanum lycopersicum L.) and its wild relatives. Int J Biol Macromol 165:2253–2266. https://doi.org/10.1016/j.ijbiomac.2020.10.080
Studer G, Biasini M, Schwede T (2014) Assessing the local structural quality of transmembrane protein models using statistical potentials (QMEANBrane). Bioinformatics 30:i505–i511. https://doi.org/10.1093/bioinformatics/btu457
Sun F, Suen PK, Zhang Y et al (2012) A dual-targeted purple acid phosphatase in Arabidopsis thaliana moderates carbon metabolism and its overexpression leads to faster plant growth and higher seed yield. New Phytol 194:206–219. https://doi.org/10.1111/j.1469-8137.2011.04026.x
Sun Q, Li J, Cheng W et al (2018) AtPAP2, a unique member of the PAP family, functions in the plasma membrane. Genes 9:257. https://doi.org/10.3390/genes9050257
Tran HT, Hurley BA, Plaxton WC (2010a) Feeding hungry plants: the role of purple acid phosphatases in phosphate nutrition. Plant Sci 179:14–27. https://doi.org/10.1016/j.plantsci.2010.04.005
Tran HT, Qian W, Hurley BA et al (2010b) Biochemical and molecular characterization of AtPAP12 and AtPAP26: the predominant purple acid phosphatase isozymes secreted by phosphate-starved Arabidopsis thaliana. Plant Cell Environ 33:1789–1803. https://doi.org/10.1111/j.1365-3040.2010.02184.x
Veljanovski V, Vanderbeld B, Knowles VL et al (2006) Biochemical and molecular characterization of AtPAP26, a vacuolar purple acid phosphatase up-regulated in phosphate-deprived Arabidopsis suspension cells and seedlings. Plant Physiol 142:1282–1293. https://doi.org/10.1104/pp.106.087171
Wang X, Wang Y, Tian J et al (2009) Overexpressing AtPAP15 enhances phosphorus efficiency in soybean. Plant Physiol 151:233–240. https://doi.org/10.1104/pp.109.138891
Wang L, Li Z, Qian W et al (2011) The Arabidopsis purple acid phosphatase AtPAP10 is predominantly associated with the root surface and plays an important role in plant tolerance to phosphate limitation. Plant Physiol 157:1283–1299. https://doi.org/10.1104/pp.111.183723
Wang L, Lu S, Zhang Y et al (2014) Comparative genetic analysis of Arabidopsis purple acid phosphatases AtPAP10, AtPAP12, and AtPAP26 provides new insights into their roles in plant adaptation to phosphate deprivation. J Integr Plant Biol 56:299–314. https://doi.org/10.1111/jipb.12184
Wiederstein M, Sippl MJ (2007) ProSA-web: interactive web service for the recognition of errors in three-dimensional structures of proteins. Nucleic Acids Res 35:W407–W410. https://doi.org/10.1093/nar/gkm290
Wu P, Ma L, Hou X et al (2003) Phosphate starvation triggers distinct alterations of genome expression in Arabidopsis roots and leaves. Plant Physiol 132:1260–1271. https://doi.org/10.1104/pp.103.021022
Yadava P, Dayaman V, Agarwal A et al (2022) Fine-tuning the transcriptional regulatory model of adaptation response to phosphate stress in maize (Zea mays L.). Physiol Mol Biol Plants 28:885–898. https://doi.org/10.1007/s12298-022-01155-x
Yang J, Zhang Y (2015) I-TASSER server: new development for protein structure and function predictions. Nucleic Acids Res 43:W174–W181. https://doi.org/10.1093/nar/gkv342
Young JC, Krysan PJ, Sussman MR (2001) Efficient screening of Arabidopsis T-DNA insertion lines using degenerate primers. Plant Physiol 125:513–518. https://doi.org/10.1104/pp.125.2.513
Zamani K, Sabet MS, Lohrasebi T et al (2012) Improved phosphate metabolism and biomass production by overexpression of AtPAP18 in tobacco. Biologia 67:713–720. https://doi.org/10.2478/s11756-012-0072-3
Zamani K, Lohrasebi T, Sabet MS et al (2014) Expression pattern and subcellular localization of Arabidopsis purple acid phosphatase AtPAP9. Gene Expr Patterns 14:9–18. https://doi.org/10.1016/j.gep.2013.08.001
Zamani K, Sabet MS, Lohrasebi T, Malboobi MA (2018) Functional analysis of AtPAP18 gene encoding a purple acid phosphatase in Arabidopsis thaliana. Mod Genet J 13:321–332
Zhang X, Henriques R, Lin S-S et al (2006) Agrobacterium-mediated transformation of Arabidopsis thaliana using the floral dip method. Nat Protoc 1:641–646. https://doi.org/10.1038/nprot.2006.97
Zhang W, Gruszewski HA, Chevone BI, Nessler CL (2008) An Arabidopsis purple acid phosphatase with phytase activity increases foliar ascorbate. Plant Physiol 146:431–440. https://doi.org/10.1104/pp.107.109934
Zhu H, Qian W, Lu X et al (2005) Expression patterns of purple acid phosphatase genes in Arabidopsis organs and functional analysis of AtPAP23 predominantly transcribed in flower. Plant Mol Biol 59:581–594. https://doi.org/10.1007/s11103-005-0183-0
Acknowledgements
This research was supported by a grant from the Tarbiat Modares University of I.R. Iran. We gratefully acknowledge the National Institute of Genetic Engineering and Biotechnology (NIGEB) for kindly supporting us in preparing of AtPAP17 construct. In the end, we should appreciate their valuable technical assistance.
Supplementary information
Table S1—Chi-square test for genotypes conformity from Mendelian 3:1 ratio (kanamycin resistant: sensitive).
Figure S1—Semi-quantitative RT-PCR test. The comparison of AtPAP17 transcript levels in (A) root or shoot of Arabidopsis WT against homozygous atpap17-mutant (Mu) line, or (B) some of distinct homozygous T3 overexpressed (OE) plants versus WT (samples prepared from whole of plant), or (C) root or shoot of WT and homozygous OE plant. RT-PCR reactions were done for 35 cycles for all experiments with the specific primers for AtPAP17 (Unique PAP17 F, R). The α-Tubulin transcript level was selected as an internal control (bottom panel). Samples were obtained by the whole of plant or shoot/root pool of WT plants, Mu line and OE plants.
Figure S2—Phenotypically comparison of 21-day-old seedlings. The AtPAP17-overexpressed (OE), atpap17-mutant (Mu) and WT plants have grown for 14 days on MS medium culture containing (A) 1.2 mM Pi (+ P) or (B) no Pi (− P). Unit scale is 1 cm.
Funding
This study was funded by a grant for a Master’s thesis from the Tarbiat Modares University of I.R. Iran (Grant No. 9085326).
Author information
Authors and Affiliations
Contributions
All the experiments, data analysis, and manuscript writing were done by AJL. MSS supervised the project and provided editorial input into the writing. MJ-J, KZ, TL, and MAM advised the project. All authors commented on previous versions of the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Jamali Langeroudi, A., Sabet, M.S., Jalali-Javaran, M. et al. Functional assessment of AtPAP17; encoding a purple acid phosphatase involved in phosphate metabolism in Arabidopsis thaliana. Biotechnol Lett 45, 719–739 (2023). https://doi.org/10.1007/s10529-023-03375-x
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10529-023-03375-x