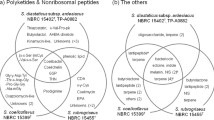
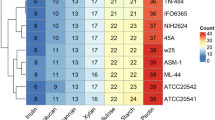
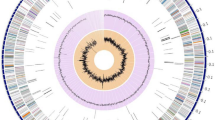
Abstract
Actinomycetes are remarkable natural sources of active natural molecules and enzymes of considerable industrial value. Streptomyces mobaraensis is the first microorganism found to produce transglutaminase with broad industrial applications. Although transglutaminase in S. mobaraensis has been well studied over the past three decades, the genome of S. mobaraensis and its secondary metabolic potential were poorly reported. Here, we presented the complete genome of S. mobaraensis DSM40587 obtained from the German Collection of Microorganisms and Cell Cultures GmbH. It contains a linear chromosome of 7,633,041 bp and a circular plasmid of 23,857 bp. The chromosome with an average GC content of 73.49% was predicted to harbour 6683 protein-coding genes, seven rRNA and 69 tRNA genes. Comparative genomic analysis reveals its meaningful genomic characterisation. A comprehensive bioinformatics investigation identifies 35 putative BGCs (biosynthesis gene clusters) involved in synthesising various secondary metabolites. Of these, 13 clusters showed high similarity (> 55%) to known BGCs coding for polyketides, nonribosomal peptides, hopene, RiPP (Ribosomally synthesized and post-translationally modified peptides), and others. Furthermore, these BGCs with over 65% similarity to the known BGCs were analysed in detail. The complete genome of S. mobaraensis DSM40587 reveals its capacity to yield diverse bioactive natural products and provides additional insights into discovering novel secondary metabolites.




Similar content being viewed by others
Abbreviations
- BGC:
-
Biosynthetic gene cluster
- RiPP:
-
Ribosomally synthesized and post-translationally modified peptide
- NRPS:
-
Non-ribosomal peptide synthetase
- PKS:
-
Polyketide synthase
- GCF:
-
Gene cluster family
- FPPS:
-
Farnesyl pyrophosphate synthetase
- GFPPS:
-
Geranylfarnesyl diphosphate synthase
References
Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, Harris MA, Hill DP, Issel-Tarver L, Kasarskis A, Lewis S, Matese JC, Richardson JE, Ringwald M, Rubin GM, Sherlock G (2000) Gene ontology: tool for the unification of biology. Nat Genet 25(1):25–29. https://doi.org/10.1038/75556
Baltz RH (2019) Natural product drug discovery in the genomic era: realities, conjectures, misconceptions, and opportunities. J Ind Microbiol Biotechnol 46(3–4):281–299. https://doi.org/10.1007/s10295-018-2115-4
Bentley SD, Chater KF, Cerdeño-Tárraga AM, Challis GL, Thomson NR, James KD, Harris DE, Quail MA, Kieser H, Harper D, Bateman A, Brown S, Chandra G, Chen CW, Collins M, Cronin A, Fraser A, Goble A, Hidalgo J, Hornsby T, Howarth S, Huang CH, Kieser T, Larke L, Murphy L, Oliver K, O’Neil S, Rabbinowitsch E, Rajandream MA, Rutherford K, Rutter S, Seeger K, Saunders D, Sharp S, Squares R, Squares S, Taylor K, Warren T, Wietzorrek A, Woodward J, Barrell BG, Parkhill J, Hopwood DA (2002) Complete genome sequence of the model actinomycete Streptomyces coelicolor A3(2). Nature 417(6885):141–147. https://doi.org/10.1038/417141a
Bérdy J (2005) Bioactive microbial metabolites. J Antibiot 58(1):1–26. https://doi.org/10.1038/ja.2005.1
Brown CA, Murray AW, Verstrepen KJ (2010) Rapid Expansion and functional divergence of subtelomeric gene families in yeasts. Curr Biol 20(10):895–903. https://doi.org/10.1016/j.cub.2010.04.027
Bu Q-T, Yu P, Wang J, Li Z-Y, Chen X-A, Mao X-M, Li Y-Q (2019) Rational construction of genome-reduced and high-efficient industrial Streptomyces chassis based on multiple comparative genomic approaches. Microb Cell Fact 18:16. https://doi.org/10.1186/s12934-019-1055-7
Bu Q-T, Li Y-P, Xie H, Wang J, Li Z-Y, Chen X-A, Mao X-M, Li Y-Q (2020) Comprehensive dissection of dispensable genomic regions in Streptomyces based on comparative analysis approach. Microb Cell Fact 19(1):99. https://doi.org/10.1186/s12934-020-01359-4
Chen Y, Zhang W, Zhu Y, Zhang Q, Tian X, Zhang S, Zhang C (2014) Elucidating hydroxylation and methylation steps tailoring piericidin A1 biosynthesis. Org Lett 16(3):736–739. https://doi.org/10.1021/ol4034176
Dahdah K, Charchar N, Bouchaala L, Nourine H, Belkabla N, Melo J, Nabti E-H (2022) Isolation, in vitro evaluation and construction of versatile microbial consortia. Cell Mol Biol (noisy-Le-Grand) 68(8):173–181. https://doi.org/10.14715/cmb/2022.68.8.31
Date M, Yokoyama K, Umezawa Y, Matsui H, Kikuchi Y (2004) High level expression of Streptomyces mobaraensis transglutaminase in Corynebacterium glutamicum using a chimeric pro-region from Streptomyces cinnamoneus transglutaminase. J Biotechnol 110(3):219–226. https://doi.org/10.1016/j.jbiotec.2004.02.011
Duarte L, Matte CR, Bizarro CV, Ayub MAZ (2019) Review transglutaminases: part II-industrial applications in food, biotechnology, textiles and leather products. World J Microbiol Biotechnol 36(1):11. https://doi.org/10.1007/s11274-019-2792-9
Duarte L, Matte CR, Bizarro CV, Ayub MAZ (2020) Transglutaminases: part I-origins, sources, and biotechnological characteristics. World J Microbiol Biotechnol 36(1):15. https://doi.org/10.1007/s11274-019-2791-x
El-Metwally M, Abdel-Mogib M, Elfedawy M, Sohsah G, Rezk A, Moustafa M, Shaaban M (2021) Isolation, purification and structure elucidation of three new bioactive secondary metabolites from Streptomyces lividans AM. Biocell. https://doi.org/10.32604/biocell.2021.013198
Emms DM, Kelly S (2019) OrthoFinder: phylogenetic orthology inference for comparative genomics. Genome Biol 20(1):238. https://doi.org/10.1186/s13059-019-1832-y
Fatima SW, Khare SK (2021) Effect of key regulators in augmenting transcriptional expression of Transglutaminase in Streptomyces mobaraensis. Bioresour Technol 340:125627. https://doi.org/10.1016/j.biortech.2021.125627
Gilchrist CLM, Chooi Y-H (2021) Clinker & clustermap. js: automatic generation of gene cluster comparison figures. Bioinformatics 37(16):2473–2475. https://doi.org/10.1093/bioinformatics/btab007
Graham-Taylor C, Kamphuis LG, Derbyshire MC (2020) A detailed in silico analysis of secondary metabolite biosynthesis clusters in the genome of the broad host range plant pathogenic fungus Sclerotinia sclerotiorum. BMC Genomics 21(1):7. https://doi.org/10.1186/s12864-019-6424-4
Gubbens J, Wu C, Zhu H, Filippov DV, Florea BI, Rigali S, Overkleeft HS, van Wezel GP (2017) Intertwined precursor supply during biosynthesis of the catecholate-hydroxamate siderophores qinichelins in Streptomyces sp. MBT76. ACS Chem Biol 12(11):2756–2766. https://doi.org/10.1021/acschembio.7b00597
Hei Y, Zhang H, Tan N, Zhou Y, Wei X, Hu C, Liu Y, Wang L, Qi J, Gao J-M (2021) Antimicrobial activity and biosynthetic potential of cultivable actinomycetes associated with Lichen symbiosis from Qinghai-Tibet Plateau. Microbiol Res 244:126652. https://doi.org/10.1016/j.micres.2020.126652
Hindra YD, Teng Q, Dong LB, Crnovčić I, Huang T, Ge H, Shen B (2017) Genome mining of Streptomyces mobaraensis DSM40847 as a bleomycin producer providing a biotechnology platform to engineer designer bleomycin analogues. Org Lett 19(6):1386–1389. https://doi.org/10.1021/acs.orglett.7b00283
Hou A, Dickschat JS (2020) The biosynthetic gene cluster for sestermobaraenes-discovery of a geranylfarnesyl diphosphate synthase and a multiproduct sesterterpene synthase from Streptomyces mobaraensis. Angew Chem Int Ed 59(45):19961–19965. https://doi.org/10.1002/anie.202010084
Hou A, Dickschat JS (2021) Using terpene synthase plasticity in catalysis: on the enzymatic conversion of synthetic farnesyl diphosphate analogues. Chemistry 27(63):15644–15649. https://doi.org/10.1002/chem.202103049
Juettner NE, Bogen JP, Bauer TA, Knapp S, Pfeifer F, Huettenhain SH, Meusinger R, Kraemer A, Fuchsbauer HL (2020) Decoding the papain inhibitor from Streptomyces mobaraensis as being hydroxylated chymostatin derivatives: purification, structure analysis, and putative biosynthetic pathway. J Nat Prod 83(10):2983–2995. https://doi.org/10.1021/acs.jnatprod.0c00201
Kang H-S, Kim E-S (2021) Recent advances in heterologous expression of natural product biosynthetic gene clusters in Streptomyces hosts. Curr Opin Biotechnol 69:118–127. https://doi.org/10.1016/j.copbio.2020.12.016
Kawata J, Naoe T, Ogasawara Y, Dairi T (2017) Biosynthesis of the carbonylmethylene structure found in the ketomemicin class of pseudotripeptides. Angew Chem Int Ed 56(8):2026–2029. https://doi.org/10.1002/anie.201611005
Kumar P, Das S, Tigga R, Pandey R, Bhattacharya SN, Taneja B (2021) Whole genome sequences of two Trichophyton indotineae clinical isolates from India emerging as threats during therapeutic treatment of dermatophytosis. 3 Biotech 11(9):402. https://doi.org/10.1007/s13205-021-02950-1
Liao C, Seebeck FP (2019) In vitro reconstitution of bacterial DMSP biosynthesis. Angew Chem Int Ed 58(11):3553–3556. https://doi.org/10.1002/anie.201814662
Liu Z, Zhang Y, Sun J, Huang W-C, Xue C, Mao X (2020) A novel soluble squalene-hopene cyclase and its application in efficient synthesis of hopene. Front Bioeng Biotechnol. https://doi.org/10.3389/fbioe.2020.00426
Liu N, Shi Y-e, Li J, Zhu M, Zhang T (2021) Identification and genome analysis of Comamonas testosteroni strain JLU460ET, a novel steroid-degrading bacterium. 3 Biotech 11(9):404. https://doi.org/10.1007/s13205-021-02949-8
Low ZJ, Pang LM, Ding Y, Cheang QW, Le Mai HK, Thi Tran H, Li J, Liu XW, Kanagasundaram Y, Yang L, Liang ZX (2018) Identification of a biosynthetic gene cluster for the polyene macrolactam sceliphrolactam in a Streptomyces strain isolated from mangrove sediment. Sci Rep 8(1):1594. https://doi.org/10.1038/s41598-018-20018-8
Mahmood KI, Najmuldeen HHR, Rachid SK (2022) Physiological regulation for enhancing biosynthesis of biofilm-inhibiting secondary metabolites in Streptomyces cellulosae: regulation for enhancing biosynthesis of antibiofilm. Cell Mol Biol (noisy-Le-Grand) 68(5):33–46. https://doi.org/10.14715/cmb/2022.68.5.5
Maxson T, Tietz JI, Hudson GA, Guo XR, Tai HC, Mitchell DA (2016) Targeting reactive carbonyls for identifying natural products and their biosynthetic origins. J Am Chem Soc 138(46):15157–15166. https://doi.org/10.1021/jacs.6b06848
Minh BQ, Schmidt HA, Chernomor O, Schrempf D, Woodhams MD, von Haeseler A, Lanfear R (2020) IQ-TREE 2: new models and efficient methods for phylogenetic inference in the genomic era. Mol Biol Evol 37(5):1530–1534. https://doi.org/10.1093/molbev/msaa015
Mosbacher TG, Bechthold A, Schulz GE (2005) Structure and function of the antibiotic resistance-mediating methyltransferase AviRb from Streptomyces viridochromogenes. J Mol Biol 345(3):535–545. https://doi.org/10.1016/j.jmb.2004.10.051
Navarro-Munoz JC, Selem-Mojica N, Mullowney MW, Kautsar SA, Tryon JH, Parkinson EI, De Los Santos ELC, Yeong M, Cruz-Morales P, Abubucker S, Roeters A, Lokhorst W, Fernandez-Guerra A, Cappelini LTD, Goering AW, Thomson RJ, Metcalf WW, Kelleher NL, Barona-Gomez F, Medema MH (2020) A computational framework to explore large-scale biosynthetic diversity. Nat Chem Biol 16(1):60–68. https://doi.org/10.1038/s41589-019-0400-9
Palaniappan N, Ayers S, Gupta S, el Habib S, Reynolds KA (2006) Production of hygromycin A analogs in Streptomyces hygroscopicus NRRL 2388 through identification and manipulation of the biosynthetic gene cluster. Chem Biol 13(7):753–764. https://doi.org/10.1016/j.chembiol.2006.05.013
Palazzotto E, Tong Y, Lee SY, Weber T (2019) Synthetic biology and metabolic engineering of actinomycetes for natural product discovery. Biotechnol Adv 37(6):107366. https://doi.org/10.1016/j.biotechadv.2019.03.005
Pasternack R, Dorsch S, Otterbach JT, Robenek IR, Wolf S, Fuchsbauer HL (1998) Bacterial pro-transglutaminase from Streptoverticillium mobaraense–purification, characterisation and sequence of the zymogen. Eur J Biochem 257(3):570–576. https://doi.org/10.1046/j.1432-1327.1998.2570570.x
Schmidt S, Adolf F, Fuchsbauer HL (2008) The transglutaminase activating metalloprotease inhibitor from Streptomyces mobaraensis is a glutamine and lysine donor substrate of the intrinsic transglutaminase. FEBS Lett 582(20):3132–3138. https://doi.org/10.1016/j.febslet.2008.07.049
Seemann T (2014) Prokka: rapid prokaryotic genome annotation. Bioinformatics 30(14):2068–2069. https://doi.org/10.1093/bioinformatics/btu153
Selvakumar D (2010) Marine Streptomyces as a novel source of bioactive substances. World J Microbiol Biotechnol 26:2123–2139. https://doi.org/10.1007/s11274-010-0415-6
Tietz JI, Schwalen CJ, Patel PS, Maxson T, Blair PM, Tai HC, Zakai UI, Mitchell DA (2017) A new genome-mining tool redefines the lasso peptide biosynthetic landscape. Nat Chem Biol 13(5):470–478. https://doi.org/10.1038/nchembio.2319
Wang Y, Tang H, Debarry JD, Tan X, Li J, Wang X, Lee TH, Jin H, Marler B, Guo H, Kissinger JC, Paterson AH (2012) MCScanX: a toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucl Acids Res 40(7):e49. https://doi.org/10.1093/nar/gkr1293
Wu R, Qin Y, Shen Q, Li P (2020) The complete genome sequence of Bacillus velezensis LPL061, an exopolysaccharide-producing bacterium. 3 Biotech 10(6):243. https://doi.org/10.1007/s13205-020-02228-y
Xia H, Zhan X, Mao XM, Li YQ (2020) The regulatory cascades of antibiotic production in Streptomyces. World J Microbiol Biotechnol 36(1):13. https://doi.org/10.1007/s11274-019-2789-4
Xu S, Xie X, Zhao Y, Shi Y, Chai A, Li L, Li B (2020) Whole-genome analysis of bacillus velezensis ZF2, a biocontrol agent that protects cucumis sativus against corynespora leaf spot diseases. 3 Biotech 10(4):186. https://doi.org/10.1007/s13205-020-2165-y
Yang Z, He J, Wei X, Ju J, Ma J (2020) Exploration and genome mining of natural products from marine Streptomyces. Appl Microbiol Biotechnol 104(1):67–76. https://doi.org/10.1007/s00253-019-10227-0
Yin X, Li Y, Zhou J, Rao S, Du G, Chen J (2021) Enhanced production of transglutaminase in Streptomyces mobaraensis through random mutagenesis and site-directed genetic modification. J Agric Food Chem. https://doi.org/10.1021/acs.jafc.1c00645
Yokoyama K, Ogaya D, Utsumi H, Suzuki M, Kashiwagi T, Suzuki E, Taguchi S (2021) Effect of introducing a disulfide bridge on the thermostability of microbial transglutaminase from Streptomyces mobaraensis. Appl Microbiol Biotechnol 105(7):2737–2745. https://doi.org/10.1007/s00253-021-11200-6
Zheng F, Zhang W, Chu X, Dai Y, Li J, Zhao H, Wen L, Yue H, Yu S (2017) Genome sequencing of strain Cellulosimicrobium sp. TH-20 with ginseng biotransformation ability. 3 Biotech 7(4):237. https://doi.org/10.1007/s13205-017-0850-2
Funding
This work was supported by the National Natural Science Foundation of China (31800031), the China Postdoctoral Science Foundation (2019M653760), Natural Science Basic Research Plan in Shaanxi Province of China (2019JQ-046), Educational Commission of Shaanxi Province (20JY009), the Specialized Scientific Research Fund Projects of Academician Shengyong Zhang (18YSZX004), and the doctoral research funding (Grant No. 17SKY013) from Shangluo University.
Author information
Authors and Affiliations
Contributions
JZQ and XLF designed and performed the experiments, and wrote the manuscript. RQZ, WGD, ZXW and JJX assisted the experiments. JW and JMG revised the manuscript. All the authors discussed the results and commented on the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Ethical Approval and consent to participate
This article does not contain data from any study with human participants or animals.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Feng, Xl., Zhang, Rq., Dong, Wg. et al. The Complete Genomic Sequence of Microbial Transglutaminase Producer, Streptomyces mobaraensis DSM40587. Biochem Genet 62, 1087–1102 (2024). https://doi.org/10.1007/s10528-023-10463-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10528-023-10463-0