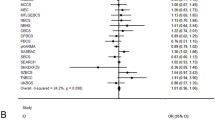
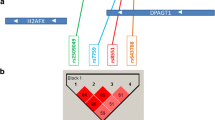
We analyzed association of potentially regulatory polymorphisms (rs590352, rs11542583, rs3829202, rs207258, and rs4796672) with breast cancer. A significant association was found between this disease and rs2072580T>A (p=0.001) located in the overlapping promoter regions of the SART3 and ISCU genes. In women with AA and AT genotypes, the risk of breast cancer is higher by 6.7 times (p=0.001) and 12 times (p=0.001), respectively, in comparison with TT genotype. Under a codominant model of inheritance (AT vs AA+TT), the risk of breast cancer was increased by 4.2 times (р=0.001) for the AT genotype. Under a recessive model of inheritance (TT vs AA+TT), the risk of disease was 10-fold higher (р=0.001) for the TT genotype. It has been demonstrated that the T>A substitution affects the binding properties of transcription factors CREB1 and REST.
Similar content being viewed by others
References
Cheng X, Wang X, Wu Z, Tan S, Zhu T, Ding K. CORO1C expression is associated with poor survival rates in gastric cancer and promotes metastasis in vitro. FEBS Open Bio. 2019;9(6):1097-1108. doi: https://doi.org/10.1002/2211-5463.12639
Coetzee SG, Coetzee GA, Hazelett DJ. motifbreakR: an R/Bioconductor package for predicting variant effects at transcription factor binding sites. Bioinformatics. 2015;31(23):3847-3849. doi: https://doi.org/10.1093/bioinformatics/btv470
Davis CA, Hitz BC, Sloan CA, Chan ET, Davidson JM, Gabdank I, Hilton JA, Jain K, Baymuradov UK, Narayanan AK, Onate KC, Graham K, Miyasato SR, Dreszer TR, Strattan JS, Jolanki O, Tanaka FY, Cherry JM. The Encyclopedia of DNA elements (ENCODE): data portal update. Nucleic Acids Res. 2018;46(D1):D794-D801. doi: https://doi.org/10.1093/nar/gkx1081
Farh KK, Marson A, Zhu J, Kleinewietfeld M, Housley WJ, Beik S, Shoresh N, Whitton H, Ryan RJ, Shishkin AA, Hatan M, Carrasco-Alfonso MJ, Mayer D, Luckey CJ, Patsopoulos NA, De Jager PL, Kuchroo VK, Epstein CB, Daly MJ, Hafler DA, Bernstein BE. Genetic and epigenetic fine mapping of causal autoimmune disease variants. Nature. 2015;518:337-43. doi: https://doi.org/10.1038/nature13835
Korbolina EE, Brusentsov II, Bryzgalov LO, Leberfarb EY, Degtyareva AO, Merkulova TI. Novel approach to functional SNPs discovery from genome-wide data reveals promising variants for colon cancer risk. Hum. Mutat. 2018;39(6):851-859. doi: https://doi.org/10.1002/humu.23425
Li J, Zhou Z, Zhang X, Zheng L, He D, Ye Y, Zhang QQ, Qi CL, He XD, Yu C, Shao CK, Qiao L, Wang L. Inflammatory Molecule, PSGL-1, Deficiency Activates Macrophages to Promote Colorectal Cancer Growth through NFκB Signaling. Mol. Cancer Res. 2017;15(4):467-477. doi: https://doi.org/10.1158/1541-7786.MCR-16-0309
Pickrell JK. Joint analysis of functional genomic data and genome-wide association studies of 18 human traits. Am. J. Hum. Genet. 2014;94(4):559-573. doi: https://doi.org/10.1016/j.ajhg.2014.03.004
Skol AD, Sasaki MM, Onel K. The genetics of breast cancer risk in the post-genome era: thoughts on study design to move past BRCA and towards clinical relevance. Breast Cancer Res. 2016 Oct 3;18(1):99.
Sveen A, Kilpinen S, Ruusulehto A, Lothe RA, Skotheim RI. Aberrant RNA splicing in cancer; expression changes and driver mutations of splicing factor genes. Oncogene. 2016; 35(19):2413-2427. doi: https://doi.org/10.1038/onc.2015.318
Ullmann P, Qureshi-Baig K, Rodriguez F, Ginolhac A, Nonnenmacher Y, Ternes D, Weiler J, Gäbler K, Bahlawane C, Hiller K, Haan S, Letellier E. Hypoxia-responsive miR-210 promotes self-renewal capacity of colon tumor-initiating cells by repressing ISCU and by inducing lactate production. Oncotarget. 2016;7(40):65454-65470. doi: https://doi.org/10.18632/oncotarget.11772
Whitmill A, Timani KA, Liu Y, He JJ. Tip110: Physical properties, primary structure, and biological functions. Life Sci. 2016;149:79-95. doi: https://doi.org/10.1016/j.lfs.2016.02.062
Zheng P, Wang W, Ji M, Zhu Q, Feng Y, Zhou F, He Q. TMEM119 promotes gastric cancer cell migration and invasion through STAT3 signaling pathway. Onco. Targets Ther. 2018;11:5835-5844. doi: https://doi.org/10.2147/OTT.S164045
Author information
Authors and Affiliations
Corresponding author
Additional information
Translated from Byulleten’ Eksperimental’noi Biologii i Meditsiny, Vol. 169, No. 1, pp. 88-91, January, 2020
Rights and permissions
About this article
Cite this article
Degtyareva, A.O., Leberfarb, E.Y., Efimova, E.G. et al. rs2072580T>A Polymorphism in the Overlapping Promoter Regions of the SART3 and ISCU Genes Associated with the Risk of Breast Cancer. Bull Exp Biol Med 169, 81–84 (2020). https://doi.org/10.1007/s10517-020-04829-2
Received:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10517-020-04829-2