Abstract
Noninvasive assessment of skin structure using hyperspectral images has been intensively studied in recent years. Due to the high computational cost of the classical methods, such as the inverse Monte Carlo (IMC), much research has been done with the aim of using machine learning (ML) methods to reduce the time required for estimating parameters. This study aims to evaluate the accuracy and the estimation speed of the ML methods for this purpose and compare them to the traditionally used inverse adding-doubling (IAD) algorithm. We trained three models – an artificial neural network (ANN), a 1D convolutional neural network (CNN), and a random forests (RF) model – to predict seven skin parameters. The models were trained on simulated data computed using the adding-doubling algorithm. To improve predictive performance, we introduced a stacked dynamic weighting (SDW) model combining the predictions of all three individually trained models. SDW model was trained by using only a handful of real-world spectra on top of the ANN, CNN and RF models that were trained using simulated data. Models were evaluated based on the estimated parameters’ mean absolute error (MAE), considering the surface inclination angle and comparing skin spectra with spectra fitted by the IAD algorithm. On simulated data, the lowest MAE was achieved by the RF model (0.0030), while the SDW model achieved the lowest MAE on in vivo measured spectra (0.0113). The shortest time to estimate parameters for a single spectrum was 93.05 μs. Results suggest that ML algorithms can produce accurate estimates of human skin optical parameters in near real-time.
Similar content being viewed by others
1 Introduction
Noninvasive, contactless optical imaging techniques have shown promise for biomedical applications, particularly in disease diagnosis, surgery, and therapy, because they are generally inexpensive, user-friendly, and provide high resolution and contrast of biological tissue.
Among these techniques, hyperspectral imaging (HSI), a hybrid optical technique that combines imaging and spectroscopy, can be used to measure spatially resolved spectra of reflected, transmitted, or fluorescent light propagating from the imaged sample to the detector. The measured data are collected in a 3D dataset known as a hypercube with two spatial dimensions (X and Y) and one spectral dimension (Z). HSI divides the light spectrum into hundreds of spectral bands covering the selected spectral region, which can include ultraviolet (UV), visible (VIS) and infrared (IR) light spectra [1, 2].
The spectrum in each image pixel results from local tissue chromophores (e.g., melanin, haemoglobin, and water) and scatterers (e.g., mitochondria, lysosomes, and collagen fibrils) [3]. It is known that the presence and progression of diseases alter spectra [4]. Based on this knowledge, HSI can provide relevant diagnostic information for the early detection of disease.
Traditionally, to obtain relevant information from hyperspectral images, biological tissue is modelled (i.e. optical properties are assigned to the tissue), and light propagation in this model is performed. The most common methods for determining optical parameters are based on simple approaches such as the Beer-Lambert (BL) law or solving the inverse scattering problem using theoretical models of light propagation in tissue [5]. The inverse diffusion approximation (IDA) is a simple and fast, but inaccurate method that applies to strongly scattering media [6, 7]. Inverse Monte Carlo is a gold standard that gives exceptionally accurate results in any geometry, but is very computationally intensive and therefore time consuming [5, 8]. Inverse adding-doubling (IAD), on the other hand, can give accurate results compared to IMC but is much faster [9]. A comparison of the GPU-accelerated IAD algorithm presented in Section 2.4 to GPU-accelerated MCML [8] confirmed that IAD was at least 2,000 times faster than MCML in extracting tissue properties from a single reflectance spectrum while achieving the absolute agreement down to the fourth decimal place. However, both approaches are too slow to enable real-time analysis of hyperspectral images or would require substantial computational resources to do so. Moreover, due to its analytic origin, IAD can only simulate light propagation in laterally infinitely layered tissue with broad homogeneous illumination.
In recent years, machine learning (ML) has been extensively studied and used to estimate various physiological parameters from diffuse reflectance spectra measurements, mainly due to its much faster fitting speed than the traditional IMC approach [10, 11]. Currently, the most widely used ML methods for analysing diffuse reflectance spectra measurements are random forests (RFs) and artificial neural networks (ANNs). In a paper by Panigrahi and Gioux, the RF algorithm is applied to the diffuse reflectance hand images, achieving an error rate of 0.556% for the absorption and 0.126% for the reduced scattering coefficient [12]. It was reported that the estimation of the parameters for a single image took 450 ms. Nguyen et al. used ANN, RF, gradient boosting machine (GBM) and generalised linear model (GLM) to extract physiological parameters from a set of simulated diffuse reflectance spectra, resulting in absolute percentage errors below 10% [11]. The models were compared with the Monte Carlo lookup table (MCLUT). ANN models outperformed all other models and achieved the best computation time. In a paper by Zhang et al., an ANN was used to estimate the scattering and absorption coefficients of human skin tissue from diffuse reflectance spectra, resulting in a relative error of 3% for the reduced scattering coefficient and 9% for the absorption coefficients [13]. Dremin et al. used neural networks on the HSI spectra in the range of 510 − 900 nm to analyse skin complications of diabetes mellitus by estimating blood volume fraction (BVF) and skin blood oxygenation (SBO) [14]. In this study, the polarisation index (PI) was used as an additional parameter to improve the model’s performance, achieving a sensitivity up to 95%. Ewerlöf et al. trained an ANN on in vivo data, which was acquired on healthy volunteers with Fitzpatrick skin type I–III [15] in order to estimate the haemoglobin oxygen saturation, achieving an average error rate of 9.5% on all evaluation data. Tsui et al. used an ANN in combination with an iterative curve-fitting algorithm to improve the speed of estimating parameters of a multi-layered skin model and reduce the computation time by 1,000 times compared to the GPU-MC approach [10]. In another paper, several ANNs were trained for different types of spatially resolved reflectance curves, achieving an average relative error of 6.1% for the absorption coefficient and 2.9% for the reduced scattering coefficient [16]. Balasubramaniam and Arnon used a cascaded feed-forward NN, where the skip connections are added to every layer, to improve image reconstruction in diffuse optical tomography achieving the improvement of 30% in contrast with the analytical solution [17]. Convolutional neural networks (CNNs) have also been shown to perform well on hyperspectral data, where most CNN architectures applied to hyperspectral data have been used for classifying different terrain types [18, 19]. In recent years, the use of CNNs is also becoming popular in medical applications. Halicek et al. proposed a CNN architecture for classifying various types of cancer from head and neck tissue samples, achieving 96.4% accuracy and outperforming support vector machines (SVMs), which achieved 92.3% accuracy [20]. Wang et al. proposed a 3D fully convolutional network called Hyper-net to segment melanoma from hyperspectral pathology images and achieved an overall accuracy of 92 % [21]. Although most CNNs are proposed for hyperspectral data to solve classification problems, there are also examples of using CNNs in regression problems. Wirkert et al. proposed a method for simulating 2D patches of multispectral images, and the 2D CNN architecture has been used to estimate oxygenation during laparoscopy [22]. Three models were compared, namely CNN, RF and BL. The error of estimating oxygenation was 12% for CNN, 13.3% for RF and 18.1% for BL, respectively.
Although much work has been done in applying ML for predictive modelling, most studies involve different imaging techniques than HSI we are presenting. For example, in the case of spatial frequency domain imaging (SFDI), information at different spatial frequencies but involving significantly fewer spectral bands is obtained. The results are maps of tissue absorption and scattering coefficients at selected spectral bands. In the case of HSI, the information about particular tissue constituents from a single spectrum (not images) or a limited number of spectral points (e.g., 6) is extracted. Our problem can be ill-posed, and therefore the tissue properties extraction performs completely different than when other imaging modalities are used. Some ML pioneering work was performed on HSI data showing that the extraction of tissue parameters is possible but the procedure was not robust [23]. However, a thorough assessment of the ML approaches to extract tissue properties from HSI data is still lacking.
For medical applications of HSI, given a large amount of hyperspectral data fast extraction of optical parameters is crucial for obtaining relevant information about biological tissue in real-time. In this study, a GPU-accelerated IAD algorithm was used for hyperspectral image analysis instead of the slower IMC method. However, IAD is still nowhere near from being applicable to analysing hyperspectral data in real-time, which inspired us to develop three different ML models based on RF, ANN and CNN. These models were trained on skin spectra simulated with a two-layer GPU-accelerated IAD algorithm and used to predict optical parameters from a dataset of 17 measured hyperspectral images of human hands acquired before, during, and after arterial occlusion. Results from all ML models were compared to IAD, and spectra calculated based on IAD and ML were compared with measured reflectance spectra. Finally, to summarise, the contributions of this work are as follows:
-
1.
We use various ML models to estimate a total of seven tissue parameters describing physiology and morphology, providing ample information about the biological tissue.
-
2.
We train all our models on a large synthetic dataset generated using the adding-doubling (AD) algorithm. This effectively eliminates the need for obtaining sizeable in vivo datasets to train complex models.
-
3.
We propose a stacked dynamic weighting (SDW) model to combine the predictions of all previously trained models by tuning it to a small number of in vivo measured diffuse reflectance spectra.
-
4.
We perform a comprehensive analysis of all models in terms of accuracy and estimation speed and show that our method can be used to rapidly extract tissue parameters (up to two images per second) from sizeable hyperspectral images, leading the way to real-time hyperspectral image analysis using GPUs and ML.
2 Materials and methods
2.1 Imaging system
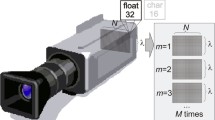
Hyperspectral images were acquired using a custom-built integrated multimodal imaging system combining a hyperspectral and an optical profilometry module presented by Rogelj et al. [24] and shown in Fig. 1. The former is a push-broom (line-scanning) hyperspectral imaging (HSI) device consisting of a 17 mm lens (Xenoplan, 1.4/47-0903, Schneider-Kreuznach, Germany), an imaging spectrograph (ImSpector V10E, Specim, Finland), a monochrome camera (Blackfly S, BFS-U3-51S5M-C, FLIR, Canada), two translation stages (8MT195, Standa, Lithuania), cross polarisers to minimise specular reflection, and a custom LED illumination panel spanning the spectral range of 400–1,000 nm. The LED light source consists of two pairs of two LED panels arranged symmetrically along the recording line to ensure uniform illumination of the sample. On the first pair of panels, white LEDs are interlaced with 780 nm LEDs; on the second pair of panels, 850 nm LEDs are interlaced with 940 nm LEDs. In total, 80 high-power LEDs are used for sample illumination. The spatial and spectral resolution of the HSI module is 200 μ m and < 1 nm, respectively. A three-dimensional (3D) optical profilometry (OP) module was also included combining a laser projector (FLEXPOINT, 30 mW, 405 nm, LASER COMPONENTS, Germany) and a monochrome camera (Flea3, FL3-U3-13Y3M-C, FLIR, Canada) with a 16 mm lens and a notch filter. 3D OP was utilised to obtain the 3D surface shape of the imaged samples and compensate for the signal loss in hyperspectral images due to sample curvature [24]. It is based on the triangulation method, where a laser projector projects a 405 nm laser line on the sample surface, and the laser line distorted due to sample surface curvature is captured by the camera from a different viewpoint. From a known relation between the camera and projector, the surface shape profile is calculated. The HSI and OP modules were calibrated using a custom-built reference object of known geometry to achieve an image misalignment of less than 0.1 mm. The accuracy of a captured 3D surface in the X, Y, and Z directions after calibration was 0.1, 0.1, and 0.05 mm, respectively.
A schematic of the multimodal imaging system combining HSI and OP [24]
2.2 Experimental procedure
Six healthy volunteers aged 23 to 24 years (three males and three females), all of whom had Caucasian skin types (Fitzpatrick types II – III), participated in this study. Each subject sat in a quiet environment and underwent a vascular occlusion test (VOT) adapted from Strömberg et al. [25]. In the experimental procedure, a blood pressure cuff was placed around the subject’s right upper arm, the hand was placed in a fixed position, and a baseline hyperspectral image of a hand was acquired along with an image of a white reflectance standard. The blood pressure cuff was then inflated to 200 mmHg and held at this level to induce arterial occlusion and prevent blood flow to the forearm and hand. After 90 seconds, when the blood oxygen level decreased dramatically, another hyperspectral image was acquired. Finally, the blood pressure cuff was removed to allow the return of oxygenated blood to the hand, and after 10 seconds a third image was acquired. For all measurements, subjects were seated in an upright position. Their hands were fixed so that the palms faced away from the camera.
In this study, 600 spectral bands were captured for each hyperspectral image in the range of 400–1,000 nm with a spectral resolution of 1 nm. Integration time was set to 200 ms and 1,000 lines were scanned for each hyperspectral image. The size of raw hypercubes was 2,048 × 1,000 × 600, resulting in more than 10 GB of data per hypercube. The total acquisition time for a single hypercube was 200 s.
The experimental protocol conforms to the principles expressed in the Declaration of Helsinki and was approved by the Slovenian National Medical Ethics Committee (0120-629/2016-3; KME 66/01/17). Informed consent was obtained from all subjects included in the study.
2.3 Image preprocessing
First, the acquired images were normalised to convert the raw hyperspectral radiance to reflectance using the following equation [1]:
where Iraw is the raw hyperspectral intensity, Iwhite is the white standard reference intensity, and Idark is the dark current measured with the camera shutter closed. In addition, image distortions due to the varying distance between the imaged object and the camera, as well as signal losses due to the changing slope of the object surface, were accounted for by using 3D data from OP and applying height and curvature corrections to hyperspectral images [24]. These corrections result in flat intensity profiles of curved objects and thus significantly reduce the effect of object curvature. In addition, hyperspectral images were spectrally reduced from the interval 400–1,000 nm to 430–750 nm with a step of 1 nm, resulting in 321 spectral bands, and spatially reduced by a factor of 10 in both X and Y directions. The wavelength range was reduced to 430–750 nm to improve the extraction of tissue parameters in the visible spectral range considered in the tissue model described in Section 2.4. Finally, image segmentation was performed to obtain a hand mask using 3D OP data and spectral angle mapper (SAM) [26]. The latter measures the spectral similarity between the spectra in each pixel of the hyperspectral image and the specified reference spectrum to distinguish the hand from the background. In our case, the reference spectrum for each image was calculated as the mean spectrum within a 20 × 20 px region of interest (ROI) in the proximal phalanx of the middle finger. As a result, the background was masked from the image, meaning only pixels inside the mask were considered in the analysis.
2.4 Image analysis
An IAD algorithm, first introduced in the field of biomedical optics by Prahl et al. [9], was developed and implemented for graphics processing units (GPUs) in MATLAB R2020b (Mathworks, MA) to enable fast and accurate simulation of light propagation in tissues. It is an iterative numerical technique for solving the radiative transport equation (RTE) in turbid media of a slab geometry. In particular, it applies to homogeneous slabs with arbitrary optical properties such as refractive index, anisotropy factor, optical thickness, albedo or phase function, taking into account the light reflection and transmission at the interface of the different layers [5, 9]. Prahl et al. [9] showed that the accuracy of the IAD is within 2–3 % of IMC if layered tissue is simulated. In our IAD algorithm, a two-layer skin model consisting of epidermis and dermis is used, and incident and outgoing light is divided into 20 fluxes to ensure reasonable accuracy so that the agreement between IAD and IMC was absolute down to the fourth decimal place.
The absorption coefficient of the epidermis used in IAD is calculated as [3, 27]:
where fm is the volume fraction of melanin,
is the melanin absorption coefficient, and
is the baseline absorption of bloodless skin. The absorption coefficient of the dermis is adopted from [3]:
where fHb and \(f_{\text {HbO}_{2}}\) are volume fractions of deoxy- and oxyhaemoglobin, μa,Hb and \(\mu _{\mathrm {a, HbO}_{2}}\) are corresponding absorption coefficients, fbrub and μa,brub are millimolar concentration and absorption coefficient of bilirubin, whereas fCO and \(f_{\text {COO}_{2}}\) are respective millimolar concentrations of reduced and oxidised cytochrome C oxidase and μa,CO and \(\mu _{\mathrm {a, COO}_{2}}\) associated absorption coefficients. Both μa,CO and \(\mu _{\mathrm {a, COO}_{2}}\) were added to the model to improve the fitting in region above 650 nm. Moreover, the reduced scattering coefficient is determined from Jacques [3] by taking into account both Mie and Rayleigh scattering:
Here, a represents the scattering coefficient at 500 nm, fRay the fraction of Rayleigh scattered light and b Mie scattering power. The refractive index of both layers was calculated as [5, 28]:
Finally, the angular distribution of scattered light intensity was given by the Henyey-Greenstein phase function [29]:
where 𝜃 is the scattering angle, and the anisotropy factor, g, was determined from Van Gemert et al. [30]:
The total number of model parameters in a two-layer skin model shown in Fig. 2 was 11, of which fm was fitted in the epidermis, fHb, \(f_{\text {HbO}_{2}}\), fbrub, fCO and \(f_{\text {COO}_{2}}\) were fitted in the dermis, and a was fitted in both layers at once. The values of fixed parameters (b, fRay, depi, and dder) are presented in Table 1, together with the accompanying references.
The calculated reflectance spectra are iteratively fitted using the Levenberg-Marquardt (LM) algorithm adopted to GPU until they match the measured values – the maximum number of iterations was set to 200.
2.5 Spectra simulation
To train the ML models, 105 spectra were simulated using the AD algorithm. Such a large number of simulated spectra provided us with enough training data and sufficient statistical accuracy to estimate the performance of different ML models. Initially, uniform sampling of the simulated parameters from the physiological ranges presented in Table 2 was implemented, where parameters b, fRay, de, and dd were fixed to values presented in Table 1.
However, this approach resulted in low ANN performance on the simulated dataset, possibly due to the combination of sampled parameters not being realistic, resulting in simulated spectra not matching the measured skin spectra. Thus, the sampling strategy was changed by assuming that the values for each parameter were sampled from a mixture of multivariate Gaussians. To fit the Gaussians, we randomly took one hyperspectral image for each phase of VOT that was later excluded from the performance evaluation and used the parameters to fit the Gaussian mixtures. Since the range of each parameter is different from the others, we first rescaled all parameters to the range of [0,1]. Then, we fitted several Gaussian mixture models (GMMs) to obtain the optimal number of clusters. All models were validated using the Silhouette score (SS) [33]. It is an internal evaluation method that determines how well the data points are clustered, considering cluster tightness and the separation between clusters. SS is calculated using the following expression:
where a(i) is the mean distance between the i-th instance and all other instances falling into the same cluster, and b(i) is the smallest mean distance from the i-th instance to all the instances not falling into the same cluster:
where d(i,j) is the distance between the instances i and j, and ∣Ci∣ is the number of instances falling into cluster i. SS takes on a value in the interval [− 1,1], where a higher value means that the clusters are tighter and well-separated, 0 indicates that the clusters are overlapping and a negative value indicates that the samples could be assigned to the wrong clusters. The clustering results shown in Fig. 3 indicate that the optimal parameter number of clusters for all cases is 2, yielding the highest SS value in all phases of the arterial occlusion test. From the spatial distribution of the parameter clusters shown in Fig. 3b, we see that the first cluster contains the parameters from the central part of the finger, which are of interest in this study, and the second cluster describes parameters from the peripheral part of the fingers considered unrepresentative of the real-world samples and treated as outliers. In these image regions, the height and curvature correction was ineffective due to the high surface inclination angle. Since the outliers deteriorate the model performance [34], we sampled the simulated points only from the first cluster (dark blue regions in Fig. 3b).
a) Silhouette score of the parameters for each phase of the arterial occlusion test (before, during, after) with respect to the number of clusters. b) The spatial distribution of the clusters is displayed on a selected hand of participant 1 recorded during VOT. The outlying parameters are shown in bright yellow, whereas the parameters of interest are shown in dark blue
The sampled parameter set was used as input to the IAD algorithm for simulating light propagation in the two-layer skin model to obtain simulated reflectance spectra before, during, and after a VOT. These spectra were eventually used to train our ML models: RF, ANN and CNN. These models were chosen due to their popularity in solving similar problems [11]. 80% of the simulated dataset was used to train the models, while the remaining 20% was used to evaluate it. Since the values for most parameters are very close to zero, using the mean absolute percentage error (MAPE) would result in large errors or even undefined results. Secondly, root mean squared error (RMSE) was not used being less robust to outliers. Therefore, we used mean absolute error (MAE), calculated by (13):
Here n represents the total number of testing instances, whereas yi and xi represent the predicted and true value of the i-th parameter, respectively.
2.6 Machine learning models
In this paper, we compared two different neural network architectures – a classical ANN with one hidden layer and a 1D CNN – and an RF model. We chose to use ANN and RF models because they are widely used in problems dealing with determining optical tissue properties [10,11,12]. Also, we use a 1D CNN model to inspect if a more complex model architecture can be more accurate compared to the simpler ones. Since the problem of determining optical tissue properties is complex, and the relation between the spectra and parameters is not linear, simple models such as linear regression were not considered in this study. All mentioned models receive single spectra as an input rather than the patches or whole hyperspectral image. Finally, we combined the predictions from all three models (ANN, 1D CNN and RF) and proposed the stacked dynamic weighting (SDW) model by training an additional ANN on a small number of in vivo measured spectra and compared it to the model that averages the predictions from all three models (AVG).
The ANN architecture used consists of three fully-connected (FC) layers. The input layer has 321 neurons, which corresponds to the dimensionality of the input spectra. The hidden layer consists of 512 neurons, using rectified linear activation units (ReLU) [35]. Different sizes of the hidden layer were examined, and the results were similar for all sizes. Finally, the output layer consists of 7 linear neurons, corresponding to the number of skin parameters.
The 1D CNN architecture shown in Fig. 4 consists of three convolutional layers followed by the max-pooling layers. All convolutional layers consist of 64 kernels of size 3 and max-pooling layers of 2. The output of the last max-pooling layer is connected to a FC layer of size 1,024, which in turn is connected to the output layer consisting of 7 neurons. Similar to the previously described ANN model, all activation functions except the output layers are ReLU, while the last layer has a linear activation function. In our experiments, we tried different sizes of FC layers and did not find any significant differences in the results.
In both neural network architectures, Adam optimiser [36] was used with a learning rate of 0.001 since a larger learning rate, such as 0.01, leads to a higher probability of deviating from the optimal solution. The size of the training batch was set to 50. Both the optimiser and the batch size were chosen empirically. The mean squared error (MSE) was used as the loss function. Input signals were scaled to [0,1], while all output parameters were normalised to a mean of 0 and a standard deviation of 1. We performed the output normalisation to ensure that the error rate for all parameters equally contributes toward the cost function. Since the measured signals usually contain noise, we augmented our simulated signals during training by adding Gaussian noise with a mean of 0 and a standard deviation of 0.007. This value of Gaussian noise was selected using the trial and error approach to maximise the performance of the models. Adding the noise also serves as a regularisation factor during training.
RF algorithm belongs to the family of ensemble learning algorithms, which are based on the idea that a single classifier/regressor is sometimes insufficient to estimate a given target class or perform a regression correctly and that multiple classifiers/regressors can be used together to improve model results [37]. Individual classifiers/regressors are highly expressive, meaning they have low bias and high variance. Because they are learned from decorrelated subsets of the training data, their joint inference equals a model having low variance while retaining low bias. In this work, we trained our RF regressor consisting of 100 trees without explicitly defining the maximum depth of each tree, inspired by the configuration used in [38]. Increasing the number of trees did not improve the results.
Finally, we built an ensemble model by combining predictions from already described models to improve the performance even more. Generally, multiple predictions can be combined using the weighted average scheme described in (14):
where fi(x) is the prediction from model i based on input spectra x, and ωi is the weight assigned to model i. Here, each ωi is constrained to range [0,1], and \({{\sum }_{1}^{n}}{\omega _{i}} = 1\). The weights can be set as static values, where the simplest approach is to weigh every model equally (AVG). However, in this study, we also explore the possibility of weighting the predictions dynamically using the architecture shown in Fig. 5 (SDW). The weighting network is a neural network consisting of one hidden layer with a ReLU activation function, receiving the predicted parameters from all three already described models together with \(\cos \limits {\phi }\) (which represents the spatial information), and outputs the model weights. The predicted parameters (inputs) are rescaled to interval [0,1] to balance out their influence. Additionally, because different physiological parameters are not necessarily correlated, it is reasonable to assume that the weights for each output parameter could differ. Therefore we train one weighting network for each parameter separately. Finally, we optimised L1 loss between the weighted predictions and the original parameters to train the weighting network. The reason for choosing L1 loss is that it is more robust to outliers that can appear in in vivo data, especially in the regions where \(\cos \limits {\phi }\) is small. The number of neurons in the hidden layer is set to 512. Experimenting with a larger hidden layer did not show any improvement. Like the already defined NN models, we used the Adam optimiser with the learning rate of 0.001, batch size set to 50, and the number of epochs set to 50. We trained weighting networks by randomly selecting three images from a single participant, one for each VOT phase. The images on which we trained our weighting network were not used in the evaluation phase.
Finally, in Fig. 6, we show the architecture of the proposed system. As can be seen, model training consists of two steps. In the first step, we generate simulated signals using the AD algorithm, which are then used to train the RF, ANN and CNN models. In the second step we perform the test of weighting networks using the single training HSI image per each VOT phase. A single weighting network is trained to predict the weights for a single parameter. After the training is done, we perform the prediction of parameters for a single spectrum using the three previously trained ML models. Then we input their predictions together with the surface inclination angle information to a weighting network. Finally, we produce the final parameter estimation using the results from the weighting network and the previously acquired predictions.
All experiments were performed on a computer consisting of two Intel®; Xeon®; Processors E5-2620 v4 CPUs, 128 GB RAM, and three GeForce RTX 2080 Ti graphics cards. Training a single ANN on one graphics card took 1.5 hours while training a CNN model took 2 hours. RF model was trained on all available CPU cores, taking 15 minutes for each parameter. Finally, the weighting networks are trained in 5 minutes per parameter since the number of real spectra is considerably smaller than the number of simulated spectra.
3 Results
3.1 Simulated data
In this section, all ML models were examined on the simulated data to demonstrate their pure performance without experimental uncertainties introduced with measured hyperspectral images. Specifically, the performance of all ML models was assessed using 10-fold cross-validation.
To begin with, Fig. 7 shows three randomly selected simulated skin spectra (IAD) and the associated skin spectra simulated from a test set of sampled parameters predicted by the three ML models. Original and predicted spectra are shown for each phase of the vascular occlusion test, namely a) before, b) during, and c) after. Visually, the predicted spectra coincide well with the simulated spectra on the entire wavelength interval. Table 3 provides the average MAE values for spectra simulated from parameters predicted by the ML models. Interestingly, the highest average spectra MAE is found for the classical ANN and the lowest for the RF model, suggesting the RF-predicted spectra match better with the IAD-simulated skin spectra than skin spectra predicted by the CNN and ANN models.
Three selected simulated skin spectra and their corresponding spectra predicted by the ML models for sampled parameters a) before, b) during, and c) after the vascular occlusion test. Original skin spectra simulated with the IAD method are shown in blue, whereas predicted spectra for ANN, CNN and RF are shown in orange, yellow and purple, respectively
Shown in Fig. 8 are the values of IAD absorption (e.g., fm, fHb, and \(f_{\text {HbO}_{2}}\)) and scattering (a) model parameters and their predictions by the three ML models. The values are predicted from the test set of the simulated skin spectra and are presented for each phase of the VOT individually. It can be seen that for the majority of parameters, the median values of predicted parameters are in good agreement with the ground truth (IAD). However, the best agreement is found for the parameters predicted by the ANN, and the worst for the parameters predicted by the RF model. The values predicted by both neural network models are less scattered than those predicted by the RF model.
Furthermore, Table 4 presents overall MAE values for all predicted parameters and vascular occlusion phases, where IAD parameters were used as ground truth for calculation. Notably, the ANN model performed best for all parameters except fCO and \(f_{\text {COO}_{2}}\), whereas the CNN model outperformed the RF model in three out of four most critical parameters (fm, \(f_{\text {HbO}_{2}}\) and a but not fHb). Train and test results in Table 4 suggest that our models do not overfit.
3.2 Experimental data
In this section, we present the results of our models on the measured data. Figure 9 shows selected spectral bands (a–c) and measured reflectance skin spectra (d–f) extracted from a hyperspectral image of the human hand of participant 2. Before the analysis, all nails were manually segmented from the hand images and omitted from the validation since the models were not trained for the nail spectra. As the surface inclination angle, ϕ, strongly affects the results, only the spectra satisfying \(\cos \limits {\phi } > 0.7\) (ϕ < 45∘) were considered. Shown in Fig. 10 is the inclination angle of the selected hand, where solid black lines outline areas satisfying the condition.
Figure 11 shows three randomly selected measured spectra (solid blue line) recorded a) before, b) during, and c) after VOT. Measured spectra are plotted with corresponding spectra fitted by the IAD algorithm (solid orange line) or predicted by ML models. Notably, spectra fitted by IAD are generally in excellent agreement with measured spectra, with slight deviations present for wavelengths above 650 nm due to higher noise levels in the measured spectra because of the LED light source properties used for imaging. The illumination intensity in that spectral region is low, and the noise is amplified. On the other hand, spectra simulated from parameters predicted by different ML models deviate more than those fitted by the IAD. Visually, the best match is found for both neural network models and the worst for the RF model, as opposed to the mean absolute error of fitted or predicted spectra with respect to measured spectra presented in Table 5.
Measured spectra (blue lines) of participant 2 recorded a) before, b) during, and c) after VOT, alongside IAD-fitted spectra (orange lines) and spectra predicted by the ANN (yellow lines), CNN (purple lines), RF (green lines), AVG (light blue lines), and SDW (dark red lines) model, respectively. Sampled spectra belong to the skin surface satisfying the condition \(\cos \limits {\phi } > 0.7\)
Shown in Fig. 12 are box plots of predicted parameters for three ML models, where IAD estimations are considered the ground truth. As can be seen, the mean parameter values of ANN and CNN are much closer to IAD than RF, yielding higher predictive accuracy. Nevertheless, the predicted values are more scattered for CNN than ANN or RF. Overall, the SDW model provides the highest prediction accuracy and precision. It is also clear that the models perform worst for fCO and \(f_{\text {COO}_{2}}\) because longer wavelengths that contain more noise than shorter wavelengths strongly affect these parameters. However, a higher noise at longer wavelengths is a system-specific problem and not a drawback of the technique itself.
To further investigate the spatial distribution of errors, the performance of all considered ML models was analysed with respect to the surface inclination angle ϕ. Specifically, the MAE of the predicted parameters was calculated, where parameters estimated by the IAD algorithm were used as the ground truth. Since the images from a single test subject were used to train the Gaussian mixture model, which was then used to generate simulated spectra, they were left out of the evaluation. The evaluation process was repeated 5 times. Each time the images from a different participant were used to train the SDW while the rest of the images were used to test the performance. The average results are shown in Fig. 13. For most parameters, the mean error of the estimated parameters decreases as the value of \(\cos \limits {\phi }\) increases. However, this is not true for \(f_{\text {COO}_{2}}\) due to the reason explained earlier in this section. Based on the data presented in Fig. 13, we can conclude that ANN and CNN are superior to RF for most parameters and VOT phases. This can also be confirmed by examining the RF box plots in Fig. 12 where especially for fbrub RF model performs much worse than the ANN and CNN models. Also, although averaging all three models does not guarantee improvement in predictive accuracy, using SDW improves the overall result. However, none of the models provided satisfactory predictions for fCO and \(f_{\text {COO}_{2}}\).
Moreover, we tested if there is a statistically significant difference in the performance of the gold standard and different ML models. Specifically, we grouped the predictions of individual parameters for all subjects and vascular occlusion phases and compared the mean values of individual parameters predicted by different methods by performing the one-way analysis of variance (ANOVA). Then, we utilised a post hoc test to determine which groups differed from each other. Generally, the results showed that predictions of individual parameters were statistically different for all models (p < 0.001). However, the post hoc test found that there was no significant difference in the mean values of fm for predictions by ANN and AVG (p = 0.624) and fHb for predictions by CNN and RF (p = 0.887) and RF and SDW (p = 0.082). There was also no difference for parameters \(f_{\text {HbO}_{2}}\) predicted by CNN and AVG (p = 0.246), fCO predicted by ANN and RF (p = 0.223), and \(f_{\text {COO}_{2}}\) predicted by CNN and SDW (p = 0.149). We can conclude that the differences in predictions of different methods used in this study are statistically significant. Nevertheless, we could not assess the performance of ML models compared to IAD based on the p-values because all values were < 0.001.
To test the time needed to estimate parameters for each algorithm, we measured the time required to estimate parameters from 105 spectra and divided the measured time with the total number of spectra to obtain the time required to estimate a single spectrum. In this test, we utilised all CPU cores for the RF and a single GPU for ANN and CNN. As seen from Table 6, ANN has the shortest estimation time because the model is the simplest and requires the least amount of arithmetic operations. On the other hand, the SDW model has the longest estimation time because it combines predictions from all three individual models, which are additionally weighted using the weighting network. AVG model takes the average prediction from all three models and is as slow as the slowest model. Also, it is crucial to notice that the RF is the slowest individual model since the individual models are used to predict each parameter. In our case, the number of pixels in the region of interest was approximately 4,500. Therefore, both weighted and individual models can estimate parameters from a hyperspectral image in less than 1.5 seconds, compared to the 27 minutes required by the IAD algorithm.
Finally, the 2D distributions of the three most crucial physiological parameters (fm, fHb and \(f_{\text {HbO}_{2}}\)) for participant 1 are shown in Fig. 14 for the IAD algorithm and all three ML models. The distribution of each parameter is visualised for three phases of the VOT. We observe that all parameter maps predicted by ML models are consistent with those extracted by the IAD method and generally feature overlapping regions of low or high parameter volume fraction.
4 Discussion
In this work, ANN, CNN, and RF models were used to estimate various physiological parameters from simulated and in vivo measured diffuse reflectance spectra. Moreover, we introduced a stacking technique, SDW, which uses an additional neural network trained on only a handful of real-world data; the network receives the predictions coupled with the spatial information and produces weights for each prediction. The models were trained with simulated spectra generated by the forward AD algorithm, and their performance was analysed on a test set of simulated spectra and measured spectra from six healthy volunteers in three phases of the vascular occlusion test (before, during, and after). Each pixel of the measured hyperspectral images was treated separately.
Initially, the performance of the three standalone ML models was tested on a subset of skin spectra simulated by the AD algorithm. These spectra served as an input to the models, whereas their output was the predicted values of seven variable optical parameters. From the predicted parameters, ML spectra were simulated using the IAD algorithm to compare their shape to the original ones.
A visual comparison of the three selected simulated skin spectra shown in Fig. 7 demonstrates a good match between the originally simulated spectra and the spectra simulated from the parameters predicted by the ML models. However, the comparison of the average MAE of spectra in Table 3 indicates that spectra simulated from values predicted by the RF model yield a lower MAE than spectra simulated from ANN and CNN predictions. This means that RF spectra match better with IAD spectra than ANN and CNN, but the average MAE is low for all models (< 0.009). We have found that MAE values are generally two to three times higher for wavelengths above 650 nm than for shorter wavelengths, primarily due to the higher noise levels in the longer wavelength region.
Moreover, box plots in Fig. 8 comparing the predicted values of parameters to the original ones (IAD) show that ANN and CNN models are superior to RF. Specifically, the median values of the former coincide with original values better than RF, and the standard deviation of predicted parameters is generally 2 to 4% lower for ANN and CNN than for RF. This is supported by the MAE values calculated for individual parameters presented in Table 4. It can be seen from this table that ANN generates a lower MAE than CNN and RF for five out of seven parameters, and CNN is superior to RF for three out of four essential parameters, as already described in Section 3.1. As a result, both neural networks can predict optical parameters from simulated spectra with higher accuracy and precision than the RF model, ANN proving slightly improved predictions compared to CNN.
Additionally, we experimented with combining the predictions from the ANN, CNN, and RF models by proposing the stacked dynamic weighting model (SDW) and comparing it against the averaged (AVG) predictions from all three models. In almost all cases, the SDW model outperforms the AVG model and all other individual models. We argue that there are two main reasons for this. First, the SDW model considers spectral inclination angle as a form of spatial information which cannot be directly simulated using AD. Second, our weighting network learned to assign smaller weights to individual models giving unreasonable or outlier predictions. This can be seen in Fig. 13 for parameter \(f_{\text {COO}_{2}}\) where the CNN clearly outperforms other individual models. In this case, our weighting network learned to prefer the CNN predictions – contrary to the AVG model, which will take the average of all individual models no matter how erroneous their predictions are.
The analysis of the results on experimental spectra provides ample insightful information. Specifically, the lowest average MAE of spectra generated from predicted model parameters was for the SDW model and the highest for the CNN, with some minor differences between all three ML models. These results are in stark contrast with the results on simulated spectra presented in Table 3, where the MAE values are generally ten times lower. Another thing to note is that the IAD algorithm, our gold standard, outperforms all ML models considerably. We hypothesise that there are two main reasons for these results. Chiefly, equivalent skin spectra can be generated using an infinite number of parameter combinations [39]. Another reason for the performance degradation of the ML algorithms is that the measurements are susceptible to noise. Although the performance of the ANN model is improved by adding Gaussian noise, the results are far from perfect. IAD seems to be less sensitive to noise due to the regularisation implemented in the ML algorithm. However, we see a significant improvement in prediction accuracy by the proposed SDW model because of the dynamic weighting of the predictions based on spatial information.
Figure 12 shows that parameter values predicted by all ML models generally agree with the ground truth (IAD). However, predictions by ANN and CNN models yield higher accuracy and are less scattered than RF model predictions. Generally, the parameters predicted by the proposed SDW model agree very well with the ground truth. We can also observe that all models perform worst for fCO and \(f_{\text {COO}_{2}}\) because experimental noise in the measured skin spectra for wavelengths larger than 650 nm strongly affects these two parameters. This could also be attributed to the high inaccuracy of the gold standard (IAD) predictions for these two parameters. We noted that high variations of fCO and \(f_{\text {COO}_{2}}\) resulted in minor changes of reflectance skin spectra, leading to non-robust predictions by the IAD algorithm, which are then also reflected in predictions by ML models. In Fig. 12, boxplots of predicted values of fCO (before, during) and \(f_{\text {COO}_{2}}\) (after) are essentially 0 due to initial parameters being set close to 0. Because the spectra shapes are not sensitive to changes in both parameters, the optimisation algorithm probably converges to a local minimum with values of fCO and \(f_{\text {COO}_{2}}\) close to zero.
We performed a one-way ANOVA test to find if the differences in the mean values of parameters predicted by different methods are statistically significant and a post hoc test to find which specific groups differed from each other. We found a statistically significant difference in parameters predicted by different methods, except in some cases presented earlier. However, we could not assess the performance of ML models compared to the gold standard based on the calculated p-values. To examine the model performance even further, in Fig. 13 we analysed how the estimation error changed with respect to the surface inclination angle ϕ and concluded that the error is highest for the surface regions where ϕ > 45∘. By observing the 2D parameter distribution maps shown in Fig. 14 for fm, fHb, and \(f_{\text {HbO}_{2}}\), and comparing the maps of ML models against the IAD, it is clear that all algorithms provide similar parameter values for specific regions of human hands.
Finally, we have shown that all ML models can provide a nearly real-time estimation speed. There are two main explanations behind this achievement. Firstly, ML models perform the parameter estimation directly on the measured hyperspectral images with the constant number of arithmetic operations per spectrum. Secondly, all models are easy to parallelise, which enables better hardware utilisation during the computation. A similar study by Nguyen et al. [11], where the estimation time of ANN and RF models was measured on CPU only, achieved the fastest time of 0.00065 s for neural networks and the slowest time of 0.67 s by querying the lookup-table. The fastest time achieved by Nguyen et al. is still up to 7 times longer than ours, suggesting that GPU additionally boosts the model performance.
As mentioned in the introduction, our work is novel in applying different ML models to estimate physiological and morphological tissue parameters directly from hyperspectral images. We demonstrated that the overall predictive performance can be improved by stacking individual ML models. Most recent research uses ML models for tissue classification or segmentation problems [20, 21]. However, the task of tissue classification and/or segmentation is simpler than regression – used in our work – and can be done satisfactorily using straightforward approaches such as spectral angle mapper SAM [2]. To their disadvantage, other research dealing with regression problems uses individual reflectance spectra, single spectral bands, or multispectral images with a much smaller number of spectral bands [10, 11, 13, 16], providing limited information about the tissue. Additionally, no other research mentioned above could estimate such a large number (7) of relevant tissue parameters as the proposed research, and hindering the disease diagnosis.
Moreover, this study uses a large spectra dataset simulated using the forward AD algorithm. As opposed to other studies that generally use the Monte Carlo (MC) technique [10, 11, 13, 16, 23], we were able to substantially increase the dataset due to a substantially shortened simulation time, improving the training of the ML models. Finally, our work demonstrated that tissue parameters could be extracted from the whole hyperspectral images in less than a second, leading the way to real-time analysis of hyperspectral images and reducing the time to establish a diagnosis.
The main limitation of our study is a small and relatively homogeneous cohort comprising six healthy volunteers aged 23 to 24 with Fitzpatrick skin types II–III. As described in Section 2.5, the seed parameters for the IAD algorithm are based on sampling the spectra from Gaussian mixture model which is learned from an in vivo measurement rather than from uniform distribution of parameters due to the poor performance of ML models. This could suggest that ML models are not robust to abnormal physical states of skin tissue, which could potentially occur in the presence of certain diseases. Therefore, considering the homogeneity of the subject pool, the IAD simulations do not represent many variations for robust testing of ML models. However, we chose to use Gaussian mixture model sampling to make the synthetic data samples used for ML model training more similar to the underlying distribution of the real-world sample. We do not believe this assumption made our experiment biased since the images used to learn the Gaussian mixture model were not taken into consideration in the test phase. On the other hand, if we used uniformly distributed sampling instead, our ML model would differ significantly from the expected real-world model, making the evaluation unrealistic because our ML model would be fitted to a different distribution than the real-world data. This study limitation could be eliminated by expanding the study cohort to include healthy subjects with various skin types and unhealthy subjects diagnosed with different skin diseases such as skin cancer, dermatitis and burns.
Several main research directions can be carried out to improve the results further. To begin with, it is possible to optimise the IAD algorithm to reduce the ill-posedness by fixing specific sets of parameters, especially those that are not interesting for skin physiology or disease diagnosis. If the fixed parameters cause too large simulation errors for the IAD algorithm, it is possible to discretise the parameters into discrete bins, converting the problem into a classification problem, which could make the method more flexible. Moreover, it is possible to analyse the variance of the IAD by estimating specific sets of parameters several times and obtaining some statistical indicators, such as the variance for each parameter. Another option is to reduce the number of spectral bands, consequently reducing the size of spectral images. Our ML models could easily be modified to input any number of spectral bands. By reducing the number of bands, models could be more robust and have better prediction accuracy and the estimation speed at the expense of spectral resolution.
From the perspective of ML, numerous possibilities can be investigated to improve the results. One potential direction would be to investigate whether the existing architectures can be improved by adding more layers or changing the cost function. To reduce the gap between the simulated and the in vivo measured spectra, one can also experiment with different autoencoder architectures or even use the state-of-the-art approach with generative adversarial networks (GANs) [40]. Finally, it would be interesting to investigate incorporating spatial information in the learning process by considering neighbouring spectra.
This paper compares different approaches to extracting tissue parameters from hyperspectral images obtained by a broad beam illumination imaging system. The approach is different from methods like SFDI, where the spatial modulation information is recorded. On the other hand, many more spectral bands (a few 100) are recorded in HSI than SFDI (a few 10). Having the additional spatial modulation information reduces ill-posedness in the case of SFDI, and thus the ML approaches are much easier to implement.
One could disregard ML models from the analysis of hyperspectral images for being too inaccurate and thus not worth the tradeoff in model development and hardware required to implement. However, with more advanced, accurate and robust ML models, ML methods will play an increasingly important role in hyperspectral image analysis, mainly due to their near real-time performance.
5 Conclusion
In this study, several ML models were used to estimate multiple physiological parameters from the simulated and in vivo measured skin reflectance spectra. The models were trained on skin spectra simulated by the IAD algorithm, which served as the ground truth, and their parameter estimation performance was analysed on a test set of simulated spectra and experimental skin spectra from hyperspectral images of hands.
The results on the simulated skin spectra show that our proposed SDW model outperforms other individual ML models in terms of yielding the lowest mean absolute error for the majority of estimated parameters. The performance of all ML models on the experimental skin spectra substantially deteriorates compared to simulated spectra. Specifically, the mean absolute error of experimental spectra is ten times higher than for simulated spectra. We also showed that the performance of all ML models is highly affected by the surface inclination angle ϕ, exhibiting that the error of estimated parameters decreases as the value of ϕ increases. Finally, we demonstrated that it is possible to predict parameters from processed hyperspectral images with ANN and CNN models in almost real-time by utilising GPUs – a hyperspectral image with approximately 4,500 pixels each can be predicted in 0.5 s.
This work proves that various ML models, such as ANN and CNN, could be trained to estimate parameters from hyperspectral images in real-time. Therefore, it is a significant step toward automating hyperspectral image analysis with the potential to use hyperspectral imaging as a real-time diagnostic tool in the clinical environment.
References
Lu G, Fei B (2014) Medical hyperspectral imaging: a review. J Biomed Opt 19(1):1–24. https://doi.org/10.1117/1.JBO.19.1.010901
Li Q, He X, Wang Y, Liu H, Xu D, Guo F (2013) Review of spectral imaging technology in biomedical engineering: achievements and challenges. J Biomed Opt 18(10):100901. https://doi.org/10.1117/1.JBO.18.10.100901
Jacques SL (2013) Optical properties of biological tissues: a review. Phys Med Biol 58(11):37–61. https://doi.org/10.1088/0031-9155/58/11/r37
Salomatina EV, Jiang B, Novak J, Yaroslavsky AN (2006) Optical properties of normal and cancerous human skin in the visible and near-infrared spectral range. J Biomed Opt 11(6):1–9. https://doi.org/10.1117/1.2398928
Bashkatov A, Genina E, Tuchin V (2011) Optical properties of skin, subcutaneous, and muscle tissues: a review. Journal of Innovative Optical Health Sciences, 04. https://doi.org/10.1142/S1793545811001319
Randeberg LL, Winnem A, Haaverstad R, Haugen OA, Svaasand LO (2005) Performance of diffusion theory vs. monte carlo methods. In: Mycek M-A (ed) Diagnostic optical spectroscopy in biomedicine III. https://doi.org/10.1117/12.633028. International Society for Optics and Photonics, vol 5862, pp 137–144
Naglič P, Vidovič L, Milanič M, Randeberg LL, Majaron B (2013) Applicability of diffusion approximation in analysis of diffuse reflectance spectra from healthy human skin. In: Spigulis J, Kuzmina I (eds) Biophotonics—Riga 2013. https://doi.org/10.1117/12.2044706. International Society for Optics and Photonics, vol 9032, pp 137–148
Wang L, Jacques SL, Zheng L (1995) Mcml—monte carlo modeling of light transport in multi-layered tissues. Comput Methods Programs Biomed 47(2):131–146. https://doi.org/10.1016/0169-2607(95)01640-F
Prahl SA, van Gemert MJC, Welch AJ (1993) Determining the optical properties of turbid media by using the adding–doubling method. Appl Opt 32(4):559–568. https://doi.org/10.1364/AO.32.000559
Tsui S, Wang C-Y, Huang T-H, Sung K (2018) Modelling spatially-resolved diffuse reflectance spectra of a multi-layered skin model by artificial neural networks trained with monte carlo simulations. Biomed Opt Express 9(4):1531– 1544
Nguyen MH, Zhang Y, Wang F, Linan JDLGE, Markey MK, Tunnell JW (2021) Machine learning to extract physiological parameters from multispectral diffuse reflectance spectroscopy. J Biomed Opt 26(5):1–10. https://doi.org/10.1117/1.JBO.26.5.052912
Panigrahi S, Gioux S (2018) Machine learning approach for rapid and accurate estimation of optical properties using spatial frequency domain imaging. J Biomed Opt 24(7):1–6. https://doi.org/10.1117/1.JBO.24.7.071606
Zhang L, Wang Z, Zhou M (2010) Determination of the optical coefficients of biological tissue by neural network. J Mod Opt 57(13):1163–1170. https://doi.org/10.1080/09500340.2010.500106
Dremin V, Marcinkevics Z, Zherebtsov E, Popov A, Grabovskis A, Kronberga H, Geldnere K, Doronin A, Meglinski I, Bykov A (2021) Skin complications of diabetes mellitus revealed by polarized hyperspectral imaging and machine learning. IEEE Trans Med Imaging 40(4):1207–1216. https://doi.org/10.1109/TMI.2021.3049591
Ewerlöf M, Strömberg T, Larsson M, Salerud EG (2022) Multispectral snapshot imaging of skin microcirculatory hemoglobin oxygen saturation using artificial neural networks trained on in vivo data. J Biomed Opt 27(3):036004
Jäger M, Foschum F, Kienle A (2013) Application of multiple artificial neural networks for the determination of the optical properties of turbid media. J Biomed Opt 18(5):1–10. https://doi.org/10.1117/1.JBO.18.5.057005
Balasubramaniam GM, Arnon S (2022) Regression-based neural network for improving image reconstruction in diffuse optical tomography. Biomed Opt Express 13(4):2006–2017. https://doi.org/10.1364/BOE.449448
Hsieh TH, Kiang JF (2020) Comparison of CNN algorithms on hyperspectral image classification in agricultural lands. Sensors (Switzerland). https://doi.org/10.3390/s20061734
Vaddi R, Manoharan P (2020) Hyperspectral image classification using CNN with spectral and spatial features integration. Infrared Physics and Technology. https://doi.org/10.1016/j.infrared.2020.103296
Halicek M, Lu G, Little JV, Wang X, Patel M, Griffith CC, El-Deiry MW, Chen AY, Fei B (2017) Deep convolutional neural networks for classifying head and neck cancer using hyperspectral imaging. Journal of Biomedical Optics. https://doi.org/10.1117/1.jbo.22.6.060503
Wang Q, Sun L, Wang Y, Zhou M, Hu M, Chen J, Wen Y, Li Q (2021) Identification of melanoma from hyperspectral pathology image using 3d convolutional networks. IEEE Trans Med Imaging 40(1):218–227. https://doi.org/10.1109/TMI.2020.3024923
Wirkert SJ (2018) Multispectral image analysis in laparoscopy – a machine learning approach to live perfusion monitoring. PhD thesis, Karlsruher Institut für Technologie (KIT. https://doi.org/10.5445/IR/1000086188
Zherebtsov E, Dremin V, Popov A, Doronin A, Kurakina D, Kirillin M, Meglinski I, Bykov A (2019) Hyperspectral imaging of human skin aided by artificial neural networks. Biomed Opt Express 10(7):3545–3559. https://doi.org/10.1364/BOE.10.003545
Rogelj L, Pavlovčič U, Stergar J, Jezeršek M., Simoncic U, Milanic M (2019) Curvature and height corrections of hyperspectral images using built-in 3d laser profilometry. Appl Opt 58:9002. https://doi.org/10.1364/AO.58.009002
Strömberg T, Sjöberg F, Bergstrand S (2017) Temporal and spatiotemporal variability in comprehensive forearm skin microcirculation assessment during occlusion protocols. Microvasc Res 113:50–55. https://doi.org/10.1016/j.mvr.2017.04.005
Kruse FA, Lefkoff AB, Boardman JW, Heidebrecht KB, Shapiro AT, Barloon PJ, Goetz AFH (1993) The spectral image processing system (sips)—interactive visualization and analysis of imaging spectrometer data. Remote Sens Environ 44(2):145–163. https://doi.org/10.1016/0034-4257(93)90013-N. Airbone Imaging Spectrometry
Jacques SL (1998) Skin optics. [Online; accessed 11-May-2021]. https://omlc.org/news/jan98/skinoptics.html Accessed 05 Nov 2021
Bashkatov AN, Genina EA, Korovina IV, Kochubey VI, Sinichkin YP, Tuchin VV (2000) In-vivo and in-vitro study of control of rat skin optical properties by action of osmotical liquid. In: Liu H, Luo Q (eds) Biomedical photonics and optoelectronic imaging. https://doi.org/10.1117/12.403935. International Society for Optics and Photonics, vol 4224, pp 300–311
Henyey LG, Greenstein JL (1940) Diffuse radiation in the Galaxy. Annales d’Astrophysique 3:117
Van Gemert MJC, Jacques SL, Sterenborg HJCM, Star WM (1989) Skin optics. IEEE Trans Biomed Eng 36(12):1146–1154. https://doi.org/10.1109/10.42108
Verdel N, Marin A, Milanič M, Majaron B (2019) Physiological and structural characterization of human skin in vivo using combined photothermal radiometry and diffuse reflectance spectroscopy. Biomed Opt Express 10(2):944–960. https://doi.org/10.1364/BOE.10.000944
Bjorgan A, Pukstad BS, Randeberg LL (2020) Hyperspectral characterization of re-epithelialization in an in vitro wound model. J Biophoton 13(10):202000108. https://doi.org/10.1002/jbio.202000108
Rousseeuw PJ (1987) Silhouettes: a graphical aid to the interpretation and validation of cluster analysis. Journal of Computational and Applied Mathematics. https://doi.org/10.1016/0377-0427(87)90125-7
Khamis A, Z I, K H, A TM (2005) The effects of outliers data on neural network performance. Journal of Applied Sciences. https://doi.org/10.3923/jas.2005.1394.1398
Nair V, Hinton GE (2010) Rectified linear units improve restricted boltzmann machines. In: Proceedings of the 27th international conference on international conference on machine learning. ICML’10. Omnipress, pp 807–814
Kingma DP, Ba J (2015) Adam: A method for stochastic optimization. In: Bengio Y, LeCun Y (eds) 3rd International Conference on Learning Representations, ICLR 2015, San Diego, CA, USA, May 7-9, 2015, Conference Track Proceedings
Breiman L (2001) Random forests. Mach Learn 45(1):5–32. https://doi.org/10.1023/A:1010933404324
Verdel N, Tanevski J, Džeroski S, Majaron B (2019) A machine-learning model for quantitative characterization of human skin using photothermal radiometry and diffuse reflectance spectroscopy. In: Choi B, Zeng H (eds) Photonics in dermatology and plastic surgery 2019. https://doi.org/10.1117/12.2509691. International Society for Optics and Photonics, vol 10851, pp 1–9
Claridge E, Hidovic-Rowe D (2014) Model based inversion for deriving maps of histological parameters characteristic of cancer from ex-vivo multispectral images of the colon. IEEE Transactions on Medical Imaging. https://doi.org/10.1109/TMI.2013.2290697
Zhu L, Chen Y, Ghamisi P, Benediktsson JA (2018) Generative adversarial networks for hyperspectral image classification. IEEE Transactions on Geoscience and Remote Sensing. https://doi.org/10.1109/TGRS.2018.2805286
Acknowledgments
The authors thank Luka Rogelj (University of Ljubljana, Faculty of Mathematics and Physics) for the hyperspectral images acquisition.
Funding
Slovenian Research Agency (ARRS) grant number P1-0389 and grant number J3-3083; Croatian Science Foundation grant number IP-2020-02-3770; University of Rijeka grant number uniri-tehnic-18-15.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of Interests
The authors declare that there are no conflicts of interest related to this article.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Manojlović, T., Tomanič, T., Štajduhar, I. et al. Rapid extraction of skin physiological parameters from hyperspectral images using machine learning. Appl Intell 53, 16519–16539 (2023). https://doi.org/10.1007/s10489-022-04327-0
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10489-022-04327-0