Abstract
The diagnosis of Crohn’s disease (CD) in the small bowel is generally performed by observing a very large number of images captured by capsule endoscopy (CE). This diagnostic technique entails a heavy workload for the specialists in terms of time spent reviewing the images. This paper presents a convolutional neural network capable of classifying the CE images to identify those ones affected by lesions indicative of the disease. The architecture of the proposed network was custom designed to solve this image classification problem. This allowed different design decisions to be made with the aim of improving its performance in terms of accuracy and processing speed compared to other state-of-the-art deep-learning-based reference architectures. The experimentation was carried out on a set of 15,972 images extracted from 31 CE videos of patients affected by CD, 7,986 of which showed lesions associated with the disease. The training, validation/selection and evaluation of the network was performed on 70%, 10% and 20% of the total images, respectively. The ROC curve obtained on the test image set has an area greater than 0.997, with points in a 95-99% sensitivity range associated with specificities of 99-96%. These figures are higher than those achieved by EfficientNet-B5, VGG-16, Xception or ResNet networks which also require an average processing time per image significantly higher than the one needed in the proposed architecture. Therefore, the network outlined in this paper is proving to be sufficiently promising to be considered for integration into tools used by specialists in their diagnosis of CD. In the sample of images analysed, the network was able to detect 99% of the images with lesions, filtering out for specialist review 96% of those with no signs of disease.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
Crohn’s disease (CD) belongs to a group of conditions known as Inflammatory Bowel Disease, characterised by chronic inflammatory processes of unknown aetiology. These processes affect primarily, although not exclusively, any area of the gastrointestinal tract. Endoscopic examinations play an important role in monitoring these patients. The majority have lesions in the terminal ileum, an area accessible by flexible endoscope (colonoscopy with ileoscopy). However, when the disease affects other areas of the small bowel (SB), patients may show unexplained clinical symptomatology and/or non-specific findings in endoscopic examinations. In this scenario, an important role is played by the endoscopy that uses video capsule or wireless capsule endoscopy, as it makes it possible to detect mucosal lesions in the proximal SB, an area inaccessible by other endoscopic techniques. The framework of this paper is the diagnosis of SB-localised CD, which can occur in up to one third of these patients [1].
Capsule endoscopy (CE) has become a frontline diagnostic procedure in the study of SB disorders [2]. Since its approval by the Food and Drugs Association (FDA) in 2001, the use of CE has improved the diagnostic imaging of SB disorders and has been recommended in clinical practice guidelines [3]. It is a non-invasive technique whereby the patient swallows a device equipped with an optical image acquisition system and battery that can capture images while, using peristaltic movements, it moves along the digestive tract. The image capturing system adapts to the transit speed of the capsule and can vary from 2 to 6 frames per second. The captured images are transmitted by radiofrequency to a belt equipped with an antenna system that the patient wears during the procedure. This belt is connected to a recording system which will store the images that will be downloaded to a computer once the study is completed. A video will then be created and viewed using a specific software provided by the capsule manufacturer.
The most important limitation of CE for the assessment of CD is that the specialist needs to spend a long time viewing the captured images, as the procedure can take up to 8-12 hours, and the capsule, as it passes through the patient’s gastrointestinal tract, captures over 60,000 digital images. At present, all these images need to be viewed and analysed by a specialist to detect inflammatory lesions in the intestinal wall which would indicate the presence of the disease.
CE can therefore be considered an effective procedure for the diagnosis of CD in the SB, but this is very labour-intensive because of the time spent viewing the videos generated by the capsules. Hence, the cost-benefit of this technique ought to be optimised by developing a technology capable of minimising the time the specialist spends analysing the images. In this context, it would be particularly important to develop tools that support the specialist by automatically analysing and screening the huge number of images involved, leaving for the specialist’s review only those images that show signs of lesions compatible with the disease.
This has been the aim of the research work outlined in this paper, i.e., a deep learning-based methodology that uses a convolutional neural network (CNN) to classify the SB images captured by CE into two categories: images with and without evidence of CD-indicating lesions. The network evaluation on a set of 7,986 images (set balanced between the two classes of our problem) has shown sensitivity and specificity levels of 99% and 96%, respectively. These values are very promising in view of integrating the methodology into specialist support tools, as they meet the requirements of such applications for their use in clinical practice: they guarantee very high levels of sensitivity in the detection of pathological images with specificity values high enough to significantly reduce the number of images that will need to be reviewed.
The rest of this paper is organised as follows: first, the state of the art on the application of CNNs for endoscopic image analysis is reviewed paying special attention to the analysis of the images captured by CE for the detection of CD (Section 2); the following section (Section 3) provides a description of the image database used in this study. Next is the description of the proposed CNN architecture and its configuration in its training and inference stages (Section 4). Then, the performance results are presented and compared with the most representative methods available in literature (Section 5). Finally, the paper ends by discussing and summarising the main conclusions that can be drawn from the work (Sections 6 and 7).
2 State of the art
From when the speed and processing power of computers has made it possible, the digitial processing of medical images has been widely used in the diagnose and detection of diseases. In recent years, the widespread use of graphics cards (GPUs) as computing units has further strengthened the development of tools based on automatic image analysis. In this context, deep learning (DL) algorithms [4, 5] are at present the most relevant technology for the analysis of different types of medical images, i.e., eye fundus [6], lung [7], cardiac [8], abdominal [9], neurological [10] and osteomuscular [11] images.
In gastroenterology, convolutional neural networks (CNNs) have been successfully employed to detect different tract-located gastrointestinal pathologies in endoscopic images, including esophagogastroduodenoscopy, colonoscopy, and capsule endoscopy images [12]. In esophagogastroduodenoscopy (EGD), CNNs have been employed for the detection of Helicobacter pylori (HP) infection [13] and gastric cancer [14]. Shichijo et al. [13] evaluated the GoogleNet CNN [15], which had been pre-trained on natural-image features through ImageNet and fine-tuned on a dataset of 32,208 images with positive or positive HP, on a test set of 11,481 images from 397 patients (72 HP positive and 325 negative). The accuracy of CNNs trained using images classified according to eight anatomical locations in the stomach can be compared with that of experienced endoscopists, but with considerable shorter diagnostic time. Hirasawa et al. [14] used the Single Shot MultiBox Detector CNN arquitecture [16] to detect gastric cancer in EGD images. They trained the network on 13,584 images, with 2,639 histologically proven gastric cancer lesions, and evaluated the diagnostic accuracy of the constructed network on an independent test set of 2,296 stomach images collected from 69 consecutive patients with 77 gastric cancer lesions. They concluded that their proposed CNN could process numerous stored endoscopic images in a very short time with a clinically relevant diagnostic ability for detecting gastric cancer. For colonoscopy, CNNs have been applied to detect and classify colorectal polyps [17, 18]. Zhang et al. [17] used a deep CNN to extract low-level features and detect and classify hyperplastic and adenomatous colorectal polyps in endoscopic images. They concluded that their proposed method can assist endoscopists to identify polyp and non-polyp images. Their proposed methodology reported similar precision as in the visual inspection by endoscopists (87.3% vs 86.4%) but higher recall and accuracy rates (87.6% versus 77.0% and 85.9% versus 74.3%, respectively). Billah and Waheed [18] applied a CNN to transfer low-level features to a Support Vector Machine (SVM) classifier, reporting accuracy of 98.34% in polyp-affected images, with a sensitivity of 98.67% and a specificity of 98.23% on standard public databases.
As for capsule endoscopy (CE), it is mainly suitable for the localisation of obscure gastrointestinal bleeding in the small bowel (SB), with a diagnostic capacity of 60-90% depending on the series [19], and the detection and follow-up of Crohn’s disease (CD) [20]. The digital processing of CE images has been used successfully to detect gastrointestinal bleeding in the SB [21,22,23,24]. The problem was initially approached by applying conventional machine learning (ML) techniques such as the extraction of vector features based on colour, textures or statistics, and the application of binary classifier, such as K-nearest neighbours, SVM or neural network [21, 22]. However, in recent years, DL algorithms have shown greater potential. Jia et al. [23] proposed the use of an 8-layer convolutional CNN with two fully connected end layers while Li et al. [24] explored the capabilities of four classical CNN architectures (LeNet [25], AlexNet [4], GoogleNet [15], and VGG-Net [26]). These models are highly accurate and suitable for the detection of bleeding. In fact, specialist-supporting software for image viewing provided by the main CE manufacturers incorporates an artificial vision module that facilitates the work of the specialist, automatically identifying images showing bleeding. However, the situation is rather different in CD. At present, all CE-captured images need to be analysed by a specialist to detect inflammatory lesions in the intestinal wall indicative of the disease. The automatic detection of these lesions is a problem that needs solving and a real challenge for the scientific community. To date, only few papers have addressed this issue. A few years ago, approaches were started using traditional ML and image analysis techniques [27, 28], and at present are being performed with CCNs. Aoki et al. [29] used a Single Shot MultiBox Detector arquitecture to detect small-bowel mucosal breaks (erosions or ulcerations). Although the results are promising, given the limited number of lesions analysed and the small size of the sample, the authors consider the study as an initial pilot study. The main limitation preventing an in-depth study and development of CNN-based algorithms to detect CD in CE images is the lack of a large set of images with a representative number of the different pathologies indicative of the disease. To date, such a database is not publicly available to the scientific community, although efforts are being made to achieve this. Thus, Vallée et al. [30] provided CrohnIPI, a CE database consisting of 3,498 images, of which 60.55% show no evidence of CD and the remaining 38.85% are pathological images with different types of CD lesions (erythema, aphtoid, oedema, ulceration, stenosis). A consensus annotation for each image was provided, obtained from the independent review of several experts. The multi-reader annotation process was described in [31]. This article highlighted the importance of generating a consensus diagnosis. It studied the influence of the quality of the annotations on the accuracy of a recurrent attention neural network, concluding that the network accuracy increases when a consensus diagnosis is used as ground-truth. Unfortunately, to date, the number of CrohnIPI images is very limited, although the authors stated that the aim was to progressively enrich it in order to further develop automatic CD lesions recognition algorithms. However, other studies have been using a private database to address the problem. Klang et al. [32] and Barash et al. [33] collected 17,640 CE images from 49 CD patients, 7,391 images with mucosal ulcers and the rest with mucosa without ulcers. Klang et al. [32] used the Xception CNN [34] to classify between normal images and images with mucosal ulcers. They demonstrated the high capacity of the network to automatically detect CE images with mucosal ulceration (they reported AUCs of 0.99 and accuracies ranging from 0.954 to 0.967). The limitation for the possible integration of the algorithms into real tools to help the specialist detect CD is that the study is focused exclusively on mucosal ulcerations, without considering other lesions compatible with the disease. As an extension of the work by [32], Barash et al. [33] applied an ordinal CNN to classify the images according to the ulcer severity, from 1 to 3. They reported overall agreement of 67% between the consensus reading and the automatic algorithm. Although they achieved high accuracy in the discrimination between grade 1 and 3 ulcerations, the performance substantially decreased when comparing grade 2 and 3 and 1 and 2 ulcerations. In this work, a specific CNN architecture was designed to address this problem of endoscopic image classification based on the presence of CD-associated lesions. In this case, the database that has been generated for the implementation of the network includes all the lesions present in the sample of images extracted from the CE videos of patients affected by the disease which were recorded for this study. Hence, the network evaluation is carried out under conditions more closely similar to those that would occur if it was applied in clinical practice.
3 Materials
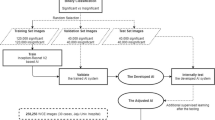
This work is part of a project funded under the 2014-2020 Andalusia ERDF Operational Programme for the implementation of a system to aid medical specialists at the Juan Ramón Jiménez hospital in Huelva (Andalusia, Spain) in the detection of CD in the SB. The work was carried out using CE videos acquired at the hospital on 31 patients affected by CD. Images were captured at 3 fps in RGB at 320x320 resolution using the PILLCAMTM SB capsule, designed and produced by Given Imaging LTD (now MEDTRONIC) [35].
Two digestive tract specialists at the hospital jointly reviewed all the video images and selected those showing lesions compatible with CD. A total of 7,986 images with lesions (pathological images) were extracted. In addition, the specialists selected an equal number of images that showed no signs of disease (healthy images). A database was thereby generated with 15,972 CE images of the SB, half of which showed signs of the disease, with clinical annotations for each image indicating the possible lesions. Table 1 shows the list of lesions observed, as well as the number and percentage of images affected by these lesions. Figure 1 shows sample images of each of these lesions.
Sample images showing the presence of CD-associated lesions in the available image set. First row: ulcers, aphthae, lymphangiectasia. Second row: bleeding, erythema, nodules. Third row: erosion, haemorrhagic suffusion, altered villi. Fourth row: atrophic mucosa, whitish villi, angiodysplasia. Fifth row: vascular lesion, multi-pathology (image with severe signs of disease; presence of different lesions in advanced stage)
The lesions showing in most of the images and affecting the largest number of patients were as follows: erosion (22 out of 31 patients), erythema (18 patients), ulcer (17), aphtha (16), bleeding (13) and lymphangiectasia (11). In addition, as shown in Table 1, there are also many images corresponding to the clinical notation “Multi-pathological”. These images, which were extracted from the videos of only 2 patients, presented many different lesions and were treated as a separate category. The remaining lesions listed in Table 1 appear in fewer than 10 patients and, although the number of images is lower and less representative than the previous ones, they were included in the image set in order to provide the most realistic scenario possible that take into account the greatest number of CD-associated lesions.
The set of 15,972 available images was divided into training, validation, and test sets, as shown in Table 2. The images were selected at random according to the percentages indicated in the table. It should be noted that the pathological images in the different subsets were selected according to the percentages of occurrence of the different lesions in the total sample of pathological images, as indicated in Table 1. As described in Section 4, the training and validation image sets were used to train and select the convolutional neural network, respectively; also, the test set was used to evaluate the selected network and to generate results (Section 5).
4 Proposed methodology
This section outlines the main aspects involved in the implementation of the convolutional neural network proposed in this work. The network was custom made for the classification of SB images into two types, “pathological” and “healthy”, based on whether or not the images show evidence of CD-compatible lesions. Its architecture is described in the first part of this section. Information is then provided on the training of the network, as well as on the criteria used for the selection of the trained network.
4.1 Network architecture
Threshold Fig. 2 is a drawing of the proposed convolutional neural network architecture. The network works on RGB images with a 320x320 resolution and consists of 6 blocks dedicated to feature extraction. Each block has a similar structure and performs the following sequence of operations:
-
Convolution. A convolution operation is performed on the input tensor with a kernel measuring 3x3, a stride of 1 and a padding of 1.
-
Batch normalisation. Batch normalisation is applied to accelerate and facilitate training convergence.
-
ReLU. The ReLU operation is applied to the batch normalisation output to introduce nonlinearities into the model.
As shown in Fig. 2, the blocks only differ in the number of convolution layers included, which progressively increase from 32 to 96.
The network design includes a pooling layer at the output of each of the above-mentioned blocks to reduce the size of the data generated. At the output of the first 5 blocks, a max-pooling operation with window size of 3x3, stride of 2 and padding of 1 is applied, whereas, at the output of the last block, global average pooling is applied.
The architecture ends with a neural network with a layer of two neurons, which are associated with each of the two classes of the problem at hand (healthy image and image affected by lesions). In addition, it should be noted that in the inference phase, the softmax function is applied to the network output. In this way, the application of the network on a given input image generates its own probability of belonging to each class.
It is worth stressing that this architecture implements an information processing strategy which is notably different from that used by the main state-of-the-art CNNs. These networks aggressively reduce the image resolution in the first layers and apply a considerably higher number of convolutions. This is because they were designed to address image classification issues of higher complexity, such as those posed by ImageNet [36], involving many different classes and images with high variability, where the elements of interest to be classified are often a relevant part of the images. These conditions greatly differ from the binary classification issue posed by this paper. Our study deals with images that present a stable environment of limited variability (internal images of the gastrointestinal tract) where the regions of interest (lesions indicative of Crohn’s disease) may be small. Therefore, it is necessary to search for an architecture tailored to our issue. In this sense, some studies [37] show that ImageNet architectures are unnecessarily large for medical imaging tasks and that it is possible to create much more efficient models in terms of parameters without a statistically significant drop in performance.
In this line, this paper suggests a network architecture resulting from an experimenting process adapted to the particular needs of our classification issue. In order to make the network sensitive to smaller lesions, a configuration has been chosen that performs most of the image processing at or close to its original resolution. However, this approach involves increasing the number of model parameters considerably. To mitigate this issue, the design contemplates using convolution layers of very small (3×3) kernels, whose number progressively increases as the layers are deeper. The architecture achieves this approach by sequentially applying blocks of similar structure (convolution operations + batch normalization + RELU) that only differ in the number of convolutions performed in each new block. Thus, convolutions are successively applied on the current resolution without disproportionately increasing the number of parameters in each block. In order to reduce the size of the data generated at the output of each block, the max-pooling operation is used, which provides certain traslation invariance and facilitates the propagation of the gradient in the first layers of the model.
4.2 Training setting
The main decisions taken in terms of the configuration of the network training process are listed below. The loss function used was cross-entropy and the optimiser chosen was stochastic gradient descent with momentum and gradient clipping by norm. The values used for the main hyperparameters were as follows:
-
Number of epochs: 2000
-
Batch size: 16
-
Learning rate: 0.0001
-
Momentum: 0.9
-
Gradient clipping by norm: 1.0
Also, the training of the network included images obtained through the application of different data augmentation operations. In particular, the following operations were applied with the probability indicated below:
-
Rotation with probability of 0.3. An angle of α ∈ (− 180, 180] was selected at random.
-
Vertical or horizontal flip with a probability of 0.5.
-
Cutout augmentation with a probability of 0.2. Between one and four regions sized between 1% and 5% of the image resolution were randomly selected.
-
Gamma contrast/gaussian noise/salt and pepper noise with a probability of 0.2. One of the three operations of reference was applied randomly.
-
Gaussian blur/random hue/random saturation with a probability of 0.2. One of the three operations was applied at random.
Figure 3 displays the images resulting from applying each operation on a sample image. The probability settings of the operations which modify the input image make it possible to maintain a significant presence of original images in the training batches. Moreover, the operations have been designed in a conservative way so that the resulting image respects the essential information of the image.
Illustration of the data augmentation operations. An original sample image is shown (first row), together with the images resulting from applying the subsequent data augmentation operations. Second row: rotation, horizontal flip, and cut-out operations. Third row: gamma contrast, gaussian and salt and pepper noise operations. Fourth row: gaussian blur, random hue, and saturation operations
4.3 Training stage and network selection
The model was trained using the training set of images, while the network selection process was based on the evaluation of the trained network at each epoch on the validation set (both sets have been referenced in Section 3 “Materials”).
Figure 4 shows the evolution of the loss function at the end of each epoch of the training process. The figure illustrates the loss value calculated by evaluating the trained network at the epoch in question in the training and validation sets.
The reference metric used to select the network was the area under the receiver operating characteristic (ROC) curve (AUC), as it allows to quantify the overall network performance at its different operating points (range of thresholds that binarise the network output probability to decide the class). Figure 5 shows the evolution of the AUC measured on the set of validation images for the different networks trained in each epoch. The maximum AUC value was reached at the 2112 epoch (AUC = 0.9953). The loss value measured in the validation set at this epoch was 0.0886 which is very close to its minimum value. Therefore, the network selected after this training process was the one using the adjusted weights after the 2112 epoch.
5 Results
The results obtained with the proposed methodology on the set of test images are presented below. Next, the results are analysed and contextualised in relation to those generated by the main state-of-the-art convolutional neural network architectures.
5.1 Proposed methodology results
The network, trained and selected according to the criteria described in the previous section, was applied on the images of this study’s test set. This generated the probability of each image being pathological (image showing CD-compatible lesions) or healthy (image with no lesions).
The metric used to quantify the overall network performance was the area under the ROC curve, AUC. The ROC curve was generated by performing a threshold sweep on the probabilities associated with the positive class which, in this case, is defined as “pathological image”. For each threshold, the network classifies the images into positive class if their probability of belonging to this class exceeds the defined threshold, or negative class (“healthy image”) if it does not. In this way, each threshold establishes a possible network operating point that can be evaluated with the binary classification metrics normally used in this type of work: accuracy (true positive and negative fraction), sensitivity (true positive fraction) or specificity (true negative fraction).
Figure 6 shows the resulting ROC curve, i.e., the representation of the true positive image fraction versus the false positive image fraction (1-specificity) associated with each threshold applied. The AUC value was 0.9978, very close to 1, which shows the network’s ability to distinguish between the images of the two classes of the problem. The network operating point with the best relation between the true and the false positive ratio –the point on the curve closest to (0,1)– was reached on a threshold of 0.37 and provides accuracy, sensitivity and specificity rates of 0.9821.
Table 3 also shows the results of these performance metrics at other representative points of the curve which were selected based on different levels of target sensitivity: 99%, 98%, 95%, and 90%. Table 4 shows the confusion matrix associated with each of these 4 selected points in the ROC curve. The target sensitivity levels were set at these values because, in this type of study, it is extremely important for the automatic image classification to have the highest possible sensitivity or, in other words, to minimise the number of false negatives, i.e., the number of images showing lesions that the network classifies as healthy. If the network was to be integrated into a specialist support tool that automatically selects images showing lesions associated with the disease, the images with lesions that were misclassified by the network would escape the review by the specialist. As shown in Table 3, the specificity percentages achieved within the established range of target sensitivities are very high, above 95% in all cases. These percentages represent the screening of healthy images that the network would perform to reduce the specialist’s workload. Thus, if for instance the proposed network was to be integrated into a real practical application configured to work at the operational point given by threshold 0.208 (sensitivity = 0.9898; specificity = 0.9630), by extrapolating the results extracted from the image sample of this study to the image set of a given CE video, it would be capable of filtering the set of images and leave to the specialist the revision of around 99% of the images with lesions and less than 4% of the images without any pathologies.
5.2 Comparison with other state-of-art CNN architectures
The proposed CNN architecture was compared with the most representative of the state-of-the-art, taking into account their proven performance in different types of medical image classification problems. The models used in this comparison were EfficientNetB5 [38], VGG-16 [26], Xception [34] and ResNet-50v2 [39].
The training configuration, as well as the selection criteria of the network to be evaluated in the set of test images, was similar to the one used for the proposed methodology, apart from the fact that these models were initialised with the weights available for the ImageNet competition [36].
Table 5 outlines the overall performance comparison of the models in the test set. As well as the AUC, the table includes the accuracy, sensitivity and specificity results at the ROC curve point where the sensitivity is closest to 0.99. As previously mentioned, this is particularly interesting from a clinical point of view, as it covers a very small number of images with lesions that are not detected by the network.
It was observed that all the networks displayed high AUC values, above 0.96 in all cases, with EfficientNetB5 and Xception, together with the CNN proposed in this work, reporting the highest performance. However, it should be pointed out that the architectures of the reference models were designed with the main objective of tackling particularly complex image classification problems involving training sets made up of millions of images and thousands of different classes (such as, for example, those proposed by ImageNet [36]). These architectures may therefore be too large to handle specific classification problems like the one at hand. This may mean that these models cannot be employed in practical application environments due to their processing time or computational capacity. Table 6 compares the models according to these issues. The models are compared in terms of the complexity of their architecture, quantified according to the number of parameters included, as well as in terms of processing speed, measured according to the average time required to process an image.
The analysis of the results provided in Table 5 concludes that the proposed convolutional neural network architecture (designed ad hoc to binary classify CE images between images with and without CD-associated lesions) provides better performance results than those obtained by state-of-the-art models. Nevertheless, its architecture is significantly less complex, resulting in much shorter processing times (see Table 6). This would not be so important if the goal was to process a small number of images. However, the main purpose of these networks is their integration into real tools that analyse videos made by capsule endoscopy. These videos generally contain a huge number of images, approximately 120,000 of them when the capsule travels 12 hours along the intestinal tract capturing images at 3fps. Therefore, as shown in Table 6, using the proposed architecture could significantly reduce the video analysis time. The most similar architectures to the one under study in terms of processing time is VGG-16 which, however, is the model with the lowest performance in terms of prediction.
6 Discussion
This paper proposed a CNN for the classification of images captured by CE to identify those showing CD-indicative lesions. The main practical application of this type of network is its integration in artificial vision tools to help specialists analyse the huge number of images included in a CE video. This is an ongoing problem that poses a major challenge to the scientific community, basically due to the difficulty in generating and having access to a large and representative set of images that includes the different lesions associated with the disease. In this context, it is important to recognise the efforts that are being made to have access to such a database. Vallée et al. in [30] created and developed CrohnIPI, a database of clinically annotated CE images. Unfortunately, there are currently only 1380 images showing the different CD-associated lesions (erythema, aphtha, oedema, ulceration, stenosis), which is insufficient to develop classification strategies based on deep learning (widely recognised as the most suitable way to address this type of problem). Thus, this line of research work needs to rely on its own generation of sets of images to implement these techniques. Hence, Klang in [32] used his own database comprising of 17,640 CE images (of which 7391 images showed signs of ulceration) in order to develop a CNN able to classify images based on the presence of this type of lesion. However, although ulcers are a major endoscopic hint for the diagnosis of the disease, their detection alone is not sufficient to develop real systems that can be used in clinical practice. The main objective of this type of support tool is to screen the images of the video, leaving those that show signs of CD for the specialist to review. This means that all lesions associated with the disease need to be taken into consideration. This was the main goal for the development of this work as there is a binary image classification problem, where the positive class corresponds to a “pathological” image affected by any lesion compatible with the disease. In this study, a total of 15,972 images were extracted from 31 CE videos of patients affected by CD, 7,986 of which showed lesions associated with the disease (Table 1 shows the different lesions that appeared in the sample, according to the clinical notes taken by two specialists in agreement, as well as their proportion in the set of images).
The CNN proposed in this work was custom designed to solve a CE image classification problem. This clearly sets it apart from the CNN models generally used in the literature to address medical image classification problems, models specially designed for the ImageNet competition (EfficientNet-B5, VGG-16, Xception and ResNet). Creating a specific network architecture made it possible to have different design choices that optimised the network performance in terms of accuracy and processing speed. The main differences between the proposed network and the other reference networks mentioned above lie in its reduced data and number of convolutions of the different layers. The networks designed for ImageNet significantly reduce the size of the data in the first layers of the network, since the relevant elements usually occupy a considerable size in the image set; moreover, the number of convolutions increases exponentially as the depth of the model expands. In contrast, the proposed network processes the data with the original size, which is particularly relevant in this case due to the small size of many of the characteristic elements of the different pathologies to be detected. Another aspect to be taken into account is the variability of the data in the images. In the case of CE images, the information is significantly more delimited than in the classification problems posed by ImageNet. Consequently, the proposed network architecture reduces the number of convolutions of its layers so that the complexity of the model is proportionate to the problem at hand. In this case study, the number of network convolutions grows linearly with increasing model depth and not exponentially as in models designed for ImageNet. Thus, this paper proposed a CNN architecture for the classification of CE images which considerably reduces the number of parameters and processing requirements compared to other state-of-the-art generic networks, without sacrificing its prediction accuracy (see Tables 5 and 6).
Regarding the results of the network on the selected set of test images, the area under the ROC curve is 0.9978. As shown in the data displayed in Table 3, the network can perform at a range of operating points that provide accuracy, sensitivity and specificity values that are adequate for the purpose for which it was designed. Therefore, the network has proved to be capable of detecting images with and without the presence of lesions, and is able to work, for example, with a sensitivity and specificity of 0.9898 and 0.9630, respectively. These values indicate that, if the network were integrated into a CD diagnostic support tool, it would allow the specialist to filter the very high number of images that make up a CE video (over 96% of the images showing no signs of lesions), leaving for review almost all images of interest showing lesions compatible with the disease (about 99%). In this regard, the analysis of false negatives (FN) (images that, although showing lesions, are predicted as “healthy” by the network and would therefore escape the specialist’s review) becomes particularly relevant from a clinical safety point of view, in terms of the potential integration of the network. Table 7 shows the distribution of the FN obtained from the corresponding operation point based on the type of lesion they show. It can be observed that the very small number of FN (only 16 out of 1597 pathological images) is largely distributed over the different types of lesions. Thus, there is no particular difficulty for the network to predict an image with a specific type of lesion as pathological. The most unfavorable case is found in images showing vascular lesions, for which the network classifies as pathological 5 out of 7 images showing this type of lesion in the test set (2 FN). This is undoubtedly related to the limited number of images available with the presence of this lesion in the training set (it is the least representative lesion in our image sample, with only 0.57% percentage of appearance, see Table 1).
To demonstrate the consistency of these results on the available images, the experiment was repeated four times following the same criteria for training, selection, and evaluation of the network: sets of training, validation and test images randomly selected with a ratio of 70, 10, and 20%, respectively (Table 2); set of pathological images created while maintaining the ratio of appearance of images with the different lesions indicative of CD (Table 1). Table 8 shows the results of this trial. The network evaluation was carried out in terms of AUC and benchmark metrics (accuracy, sensitivity, and specificity) measured at a representative point on the curve (the selection fell on the one closest to the target sensitivity of 99%). The mean AUC values, as well as the accuracy, sensitivity, and specificity values measured at the selected operating point, accompanied by the reduced standard deviation values obtained, confirm the network’s ability and adequacy to discriminate between the two types of images under study.
7 Conclusions
The main contributions of this work can be summarized as follows:
-
The presentation of a convolutional neural network designed to classify endoscopic images showing lesions associated with Crohn’s disease. The results achieved on a balanced test set of 3,194 images are promising enough to propose the integration of the network into tools that will support specialists in the diagnosis of this disease. The measured mean AUC was 0.9973 and the network is capable of functioning at an operation point able to detect 99% of the images with lesions, filtering out for specialist review 95% of images with no signs of the disease.
-
In terms of prediction accuracy and processing speed, the architecture of the proposed network proves to be more efficient than other state-of-the-art reference architectures (EfficientNet-B5, VGG-16, Xception or ResNet) widely used in medical image classification problems. The closest case is the Xception network used in Klang [32] for the classification of endoscopic images depending on whether they show mucosal ulcerations. This network, together with the one proposed in this work, are the best performers in terms of AUC (0.9976 vs. 0.9978). However, Xception’s processing speed is significantly slower, taking more than twice as long to process an image (18 ms vs. 8 ms), a difference that becomes even more relevant considering that the aim is the analysis of a CE video comprising of a vast number of images. It can therefore be concluded that it is important to focus on network models specifically designed and adapted to the problem at hand. This will guarantee, or even improve, as in this case, the accuracy of the main networks available in the literature with greater computational efficiency.
However, the main limitation of this study is the small number of image samples used for training and evaluating the network. Because of this, it was decided to generate training, validation, and test sets reflecting the real occurrence of the different CD-associated injuries (based on our sample of 15,972 images taken from 31 CE videos). Given the small number of videos/patients available, not all types of lesions are sufficiently represented in our sample of images. Therefore, to generate the training, validation, and test sets, all available videos had to be used in order to maximise the representativeness of all lesions while maintaining their occurrence proportion. For this reason, more data need to be added to these sets. Unfortunately, this is a slow and costly process that, in this study, is taking place in real time as tests are performed on new patients affected by the disease. To this effect, the planned line of action for the integration of the network in real clinical practice at the hospital where all the tests have been performed, can be summarised as follows:
-
1.
Collection of new CE videos of Crohn’s disease patients to enrich the image sets available for network training.
-
2.
Evaluation of the network at video/patient level. This will allow a more realistic quantification of the network performance in detecting images with lesions.
-
3.
Development of a second network that classifies, at lesion level, the pathological images detected by the proposed network.
-
4.
Implementation of software that will read and process CE videos and that will integrate the aforementioned networks and analyse the images so that the specialist will only review those classified as pathological. Such software must be fully validated by using it in parallel with the current programmes used by specialists to analyse all the images included in a CE video.
References
Rameshshanker R, Arebi N (2012) Endoscopy in inflammatory bowel disease when and why. World J Gastrointest Endosc 4(6):201
Wanq A, Banerjee S, Barth B, Baht Y, Chauhan S, Gottlieb S (2013) Wireless capsule endoscopy. technology status evaluation report. Gastrointest Endosc 78:805–815
Ladas S, Triantafyllou K, Spada C, Riccioni ME, Rey J-F, Niv Y, Delvaux M, De Franchis R, Costamagna G (2010) European society of gastrointestinal endoscopy (esge): recommendations (2009) on clinical use of video capsule endoscopy to investigate small-bowel, esophageal and colonic diseases. Endoscopy 42(03):220–227
Krizhevsky A, Sutskever I, Hinton GE (2012) Imagenet classification with deep convolutional neural networks. Adv Neural Inf Process Syst 25:1097–1105
LeCun Y, Bengio Y, Hinton G (2015) Deep learning. Nature 521(7553):436–444
Gulshan V, Peng L, Coram M, Stumpe MC, Wu D, Narayanaswamy A, Venugopalan S, Widner K, Madams T, Cuadros J et al (2016) Development and validation of a deep learning algorithm for detection of diabetic retinopathy in retinal fundus photographs. Jama 316(22):2402–2410
Cicero M, Bilbily A, Colak E, Dowdell T, Gray B, Perampaladas K, Barfett J (2017) Training and validating a deep convolutional neural network for computer-aided detection and classification of abnormalities on frontal chest radiographs. Investig Radiol 52(5):281–287
Ngo TA, Lu Z, Carneiro G (2017) Combining deep learning and level set for the automated segmentation of the left ventricle of the heart from cardiac cine magnetic resonance. Med Image Anal 35:159–171
Lu F, Wu F, Hu P, Peng Z, Kong D (2017) Automatic 3d liver location and segmentation via convolutional neural network and graph cut. Int J Comput Assist Radiol Surg 12(2):171–182
van der Burgh HK, Schmidt R, Westeneng H-J, de Reus MA, van den Berg LH, van den Heuvel MP (2017) Deep learning predictions of survival based on mri in amyotrophic lateral sclerosis. NeuroImage: Clin 13:361–369
Forsberg D, Sjöblom E, Sunshine JL (2017) Detection and labeling of vertebrae in mr images using deep learning with clinical annotations as training data. J Digit Imaging 30(4):406–412
Min JK, Kwak MS, Cha JM (2019) Overview of deep learning in gastrointestinal endoscopy. Gut Liver 13(4):388
Shichijo S, Nomura S, Aoyama K, Nishikawa Y, Miura M, Shinagawa T, Takiyama H, Tanimoto T, Ishihara S, Matsuo K et al (2017) Application of convolutional neural networks in the diagnosis of helicobacter pylori infection based on endoscopic images. EBioMedicine 25:106–111
Hirasawa T, Aoyama K, Tanimoto T, Ishihara S, Shichijo S, Ozawa T, Ohnishi T, Fujishiro M, Matsuo K, Fujisaki J et al (2018) Application of artificial intelligence using a convolutional neural network for detecting gastric cancer in endoscopic images. Gastric Cancer 21(4):653–660
Szegedy C, Liu W, Jia Y, Sermanet P, Reed S, Anguelov D, Erhan D, Vanhoucke V, Rabinovich A (2015) Going deeper with convolutions. In: proceedings of the IEEE conference on computer vision and pattern recognition, pp 1–9
Liu W, Anguelov D, Erhan D, Szegedy C, Reed S, Fu C-Y, Berg AC (2016) Ssd: Single shot multibox detector. In: European conference on computer vision. Springer, pp 21–37
Zhang R, Zheng Y, Mak TWC, Yu R, Wong SH, Lau JY, Poon CC (2016) Automatic detection and classification of colorectal polyps by transferring low-level cnn features from nonmedical domain. IEEE J Biomed Health Inform 21(1):41–47
Billah M, Waheed S (2018) Gastrointestinal polyp detection in endoscopic images using an improved feature extraction method. Biomed Eng Lett 8(1):69–75
Leighton JA, Triester SL, Sharma VK (2006) Capsule endoscopy: a meta-analysis for use with obscure gastrointestinal bleeding and crohn’s disease. Gastrointest Endosc Clin 16(2):229–250
Swain P (2005) Wireless capsule endoscopy and crohn’s disease. Gut 54(3):323–326
Ghosh T, Bashar SK, Fattah SA, Shahnaz C, Wahid KA (2014) A feature extraction scheme from region of interest of wireless capsule endoscopy images for automatic bleeding detection. In: 2014 IEEE international symposium on signal processing and information technology (ISSPIT). IEEE, pp 000256–000260
Xiong Y, Zhu Y, Pang Z, Ma Y, Chen D, Wang X (2015) Bleeding detection in wireless capsule endoscopy based on mst clustering and svm. In: 2015 IEEE workshop on signal processing systems (SiPS). IEEE, pp 1–4
Jia X, Meng MQ-H (2016) A deep convolutional neural network for bleeding detection in wireless capsule endoscopy images. In: 2016 38th annual international conference of the IEEE engineering in medicine and biology society (EMBC). IEEE, pp 639–642
Li P, Li Z, Gao F, Wan L, Yu J (2017) Convolutional neural networks for intestinal hemorrhage detection in wireless capsule endoscopy images. In: 2017 IEEE international conference on multimedia and expo (ICME). IEEE, pp 1518–1523
LeCun Y, Bottou L, Bengio Y, Haffner P (1998) Gradient-based learning applied to document recognition. Proc IEEE 86(11):2278–2324
Simonyan K, Zisserman A (2014) Very deep convolutional networks for large-scale image recognition. arXiv:1409.1556
Kumar R, Zhao Q, Seshamani S, Mullin G, Hager G, Dassopoulos T (2011) Assessment of crohn’s disease lesions in wireless capsule endoscopy images. IEEE Trans Biomed Eng 59(2):355–362
Jebarani WSL, Daisy VJ (2013) Assessment of crohn’s disease lesions in wireless capsule endoscopy images using svm based classification. In: 2013 international conference on signal processing, image processing & pattern recognition. IEEE, pp 303–307
Aoki T, Yamada A, Aoyama K, Saito H, Fujisawa G, Odawara N, Kondo R, Tsuboi A, Ishibashi R, Nakada A et al (2020) Clinical usefulness of a deep learning-based system as the first screening on small-bowel capsule endoscopy reading. Dig Endosc 32(4):585–591
Vallée R, De Maissin A, Coutrot A, Mouchère H, Bourreille A, Normand N (2020) Crohnipi: An endoscopic image database for the evaluation of automatic crohn’s disease lesions recognition algorithms. In: medical imaging 2020: biomedical applications in molecular, structural, and functional imaging, vol 11317. International Society for Optics and Photonics, p 113171
de Maissin A, Vallée R, Flamant M, Fondain-Bossiere M, Le Berre C, Coutrot A, Normand N, Mouchère H, Coudol S, Trang C et al (2021) Multi-expert annotation of crohn’s disease images of the small bowel for automatic detection using a convolutional recurrent attention neural network. Endosc Int Open 9(07):1136–1144
Klang E, Barash Y, Margalit RY, Soffer S, Shimon O, Albshesh A, Ben-Horin S, Amitai M, Eliakim R, Kopylov U (2020) Deep learning algorithms for automated detection of crohn’s disease ulcers by video capsule endoscopy. Gastrointest endosc 91(3):606–613
Barash Y, Azaria L, Soffer S, Yehuda RM, Shlomi O, Ben-Horin S, Eliakim R, Klang E, Kopylov U (2021) Ulcer severity grading in video capsule images of patients with crohn’s disease: an ordinal neural network solution. Gastrointest Endosc 93(1):187– 192
Chollet F (2017) Xception: deep learning with depthwise separable convolutions. In: proceedings of the IEEE conference on computer vision and pattern recognition, pp 1251–1258
MEDTRONIC: PILCAM SB SYSTEM. https://www.medtronic.com/covidien/en-us/products/capsule-endoscopy/pillcam-sb-3-system.html. Accessed 22 Dec 2021
Deng J, Dong W, Socher R, Li L-J, Li K, Fei-Fei L (2009) Imagenet: a large-scale hierarchical image database. In: 2009 IEEE conference on computer vision and pattern recognition. IEEE, pp 248–255
Ke A, Ellsworth W, Banerjee O, Ng AY, Rajpurkar P (2021) Chextransfer: performance and parameter efficiency of imagenet models for chest x-ray interpretation. In: proceedings of the conference on health, inference, and learning, pp 116–124
Tan M, Le QV (2019) Efficientnet: rethinking model scaling for convolutional neural networks. arXiv:1905.11946
He K, Zhang X, Ren S, Sun J (2016) Identity mappings in deep residual networks. In: European conference on computer vision. Springer, pp 630–645
Acknowledgements
This work was part of a project funded under the 2014-2020 Andalusia ERDF Operational Programme (Project Reference: UHU-1257810 - PO FEDER 2014-2020)
Funding
Open Access funding provided thanks to the CRUE-CSIC agreement with Springer Nature.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of Interests
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Marin-Santos, D., Contreras-Fernandez, J.A., Perez-Borrero, I. et al. Automatic detection of crohn disease in wireless capsule endoscopic images using a deep convolutional neural network. Appl Intell 53, 12632–12646 (2023). https://doi.org/10.1007/s10489-022-04146-3
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10489-022-04146-3