Abstract
Traumatic Brain Injury (TBI) could lead to intracranial hemorrhage (ICH), which has now been identified as a major cause of death after trauma if it is not adequately diagnosed and properly treated within the first 24 hours. CT examination is widely preferred for urgent ICH diagnosis, which enables the fast identification and detection of ICH regions. However, the use of it requires the clinical interpretation by experts to identify the subtypes of ICH. Besides, it is unable to provide the details needed to conduct quantitative assessment, such as the volume and thickness of hemorrhagic lesions, which may have prognostic importance to the decision-making on emergency treatment. In this paper, an optimal deep learning framework is proposed to assist the quantitative assessment for ICH diagnosis and the accurate detection of different subtypes of ICH through head CT scan. Firstly, the format of raw input data is converted from 3D DICOM to NIfTI. Secondly, a pre-trained multi-class semantic segmentation model is applied to each slice of CT images, so as to obtain a precise 3D mask of the whole ICH region. Thirdly, a fine-tuned classification neural network is employed to extract the key features from the raw input data and identify the subtypes of ICH. Finally, a quantitative assessment algorithm is adopted to automatically measure both thickness and volume via the 3D shape mask combined with the output probabilities of the classification network. The results of our extensive experiments demonstrate the effectiveness of the proposed framework where the average accuracy of 96.21 percent is achieved for three types of hemorrhage. The capability of our optimal classification model to distinguish between different types of lesion plays a significant role in reducing the false-positive rate in the existing work. Furthermore, the results suggest that our automatic quantitative assessment algorithm is effective in providing clinically relevant quantification in terms of volume and thickness. It is more important than the qualitative assessment conducted through visual inspection to the decision-making on emergency surgical treatment.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
Traumatic Brain Injury (TBI) is a sort of head injury that causes high mortality and physical disability worldwide [1]. The extreme cases meeting the recommended criteria for surgery require urgent medical and surgical management. For this reason, an accurate and prompt diagnosis is essential for the effective treatment carried out by a medical professional.
Currently, computed Tomography (CT) is accepted as one of the most common techniques applied for preliminary examination before the start of any operative procedures. It provides a low-cost solution for doctors to diagnose TBI [2]. The doctor can receive more information about patients from CT during diagnosis, follow-up, and decision-making on surgery [3]. CT is relied on to identify different diseases such as bony defects [4, 5], lung cancer [6, 7], sports-induced injuries [8], and COVID-19 [9,10,11]. CT provides a means of rapid examination for analyzing the TBI in patients [12, 13]. It also allows doctors to detect hemorrhagic lesions and determine whether immediate surgery is required for the patients [14].
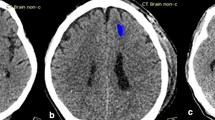
The recommended criteria for surgical consideration are detailed in [15, 16]. The patients will be considered to need surgery, for example, when the thickness of epidural hematoma (EDH) exceeds 15 mm, the thickness of subdural hematoma (SDH) reaches above 10 mm, or the lesion volume of intraparenchymal hematoma (IPH) exceeds 50 mL. It is worth noting that three out of five subtypes of intracranial hemorrhage (ICH), including EDH, SDH, and IPH, are related under the surgical consideration criteria. Additionally, the thickness of extra-axial hemorrhage (EDH and SDH) and the volume of IPH are significant to surgical consideration. These three subtypes can be distinguished by their shape and position. Figure 1 shows the images of healthy brain and those indicating these three subtypes of hemorrhage. As shown in Fig. 1b, EDH represents a biconvex shape of bleeding which occurs between the dura and skull. Distinct from EDH, SDH is a collection of blood that shows a concave shape. It exists between the dura and arachnoid mater as shown in Fig. 1c. Even though both EDH and SDH can be observed in different layers of potential space outside the brain, their position is often adjacent to the skull area on CT scan. Figure 1d demonstrates the hemorrhagic regions of IPH which is observable in the area of brain parenchyma on CT scan. However, the shape of IPH is irregular. In order to estimate the volume of IPH, it is assumed in this paper that IPH has a spherical shape.
The measurement of hemorrhagic lesions for their volume and thickness is often challenging due to the possibility that different types of hemorrhage can appear on the same CT scan. In order to estimate the thickness and volume of hemorrhagic lesions, it is necessary for the radiologists to know the subtype of each hemorrhage lesion. On this basis, the thickness and volume of each hemorrhage lesion are measured separately according to the exact subtype. In this study, the thickness of EDH and SDH is measured while the volume of IPH is calculated. However, it takes plenty of time to perform the manual measurement of thickness and volume as well as the segmentation of lesions.
Furthermore, due to the shortage of radiologists and other medical practitioners in some places such as those small hospitals in rural areas, additional techniques or tools are required to solve these problems. Rapid ICH diagnosis can help significantly reduce the death rate and boost the chances of survival for patients. This is essential for improving clinical outcome for patients. In this sense, it is practically significant to develop an intelligent algorithm that can be applied to detect different lesion types accurately and efficiently and to quantify the size of lesion for the early diagnosis of ICH.
According to literature review, there has been little attention paid to exploring the method used to estimate both thickness and volume for various subtypes of brain hemorrhage. Computer lacks the ability to follow the same diagnostic process as humans. A possible technique that can help the computer measure the thickness and volume of different hemorrhage subtypes is to evaluate their contour sizes separately. Nonetheless, there are quite few approaches to brain hemorrhage segmentation that are appropriate for different subtypes of hemorrhagic region. In this study, a framework is proposed to measure both thickness and volume of each subtype of hemorrhagic lesion (EDH, SDH, and IPH) automatically. The main contributions of this paper are summarized as follows. On the one hand: a novel method is proposed to classify and segment different types of traumatic brain injury by integrating the features extracted from a double-branch deep neural network. This network consists of a modified transfer-learning enhanced-based multi-label classifier and an optimal multi-class segmentation algorithm. On the other hand, a new algorithm of quantitative assessment is put forward to measure the thickness and volume of three-dimensional (3D) head CT scans, while each 3D scan is a stack of many 2D images called slices. The proposed algorithm can help radiologists with diagnosis and decision-marking on emergency surgery.
2 Related works
This section presents a review of the relevant literature. Then, a summary is made of all the previous studies on the methods of classification and segmentation for traumatic brain injury.
2.1 Classification model for traumatic brain injury
Over the past few years, such prominent machine learning methods as decision tree [17] and support vector machine [18, 19] have been proposed to detect hemorrhages. Despite the high accuracy achieved by most of these methods, they are still limited to detecting intracerebral hemorrhage which is also known as intraparenchymal hemorrhage (IPH). In some recent studies, deep learning methods have been mentioned for the automatic detection of multiple types of hemorrhages [20]. Jnawali et al. [21] constructed the ensemble networks with three 3D convolutional neural networks (CNNs) for the detection of intracranial hemorrhage. Lee et al. [22] trained an ensemble model consisting of four deep convolutional neural networks (DCNNs) for small datasets. Burduja et al. [23] put forward a hemorrhage detection system through the design of a lightweight CNN with long short-term memory (LSTM). Additionally, there were other CNN-LSTM models [24, 25] proposed. He [26] combined the results of SE-ResNetXt50 and EfficientNet-B3 deep neural network architecture to detect the intracranial hemorrhage and its subtypes on head CT scans. Though these models can help detect hemorrhage accurately, this model is incapable to determine the location and size of ICH.
2.2 Segmentation method for traumatic brain injury
Convolutional Neural Networks (CNNs) can produce outstanding performance in performing various tasks related to computer vision such as vehicle recognition [27, 28], image generation [29, 30], and the segmentation of automatic hemorrhagic lesion on CT scans. Farzaneh et al. [31] proposed an approach to SDH segmentation for TBI using a conventional feature extraction algorithm and a TreeBagger classifier. Remedios et al. [32] created the U-Net with transferred weight as multisite learning models (MSL). Hssayeni et al. [33] constructed a deep fully convolutional network (FCN) to segment the ICH regions on CT scans. Although the prior studies have produced impressive results in segmenting the hemorrhagic lesions of different types, there remains a problem that arises from segmenting different types of hemorrhage on the same CT slice. To solve the problem of multi-class segmentation, Kuo et al. [34] proposed the application of patch-based fully convolutional neural network (PatchFCN) for acute intracranial hemorrhage on head CT. Though the PatchFCN provided the evaluation metrics of classification with pixel-level supervision, the quantitative evaluation of various lesions was ignored. Monteiro et al. [35] worked out the design of automatic segmentation for head CT lesions system with DeepMedic [36] backbone and data augmentation. DeepMedic is a widely-known dual pathway 3D CNN architecture intended for the task of medical image segmentation. Although PatchFCN and DeepMedic can make distinction between different types of hemorrhagic lesion, it remains necessary for experts to estimate the size of lesions. Monteiro et al. [37] demonstrated the capability of a CNN through the multi-class lesion quantification and detection. This study contributed to the multi-class lesion segmentation and volume evaluation of each hemorrhagic subtype. Nonetheless, the method provided the contour of EDH and SDH in the same group as extra-axial hemorrhage (EAH) and required the involvement of experts in assessing the quantitative information of hemorrhage.
2.3 Automatic quantitative information calculation for multiple subtypes of ICH
The factors that can influence medical diagnosis and surgical consideration include hemorrhage position, hemorrhage volume, surgical timing, and curative effect [38]. In order to determine the volume of acute ICH lesions automatically, Jain et al. [39] proposed an automated image analysis based on an extension to U-Net model called icobrain which can compute the volume and midline shift of acute intracranial lesions. However, it can identify only one category of hemorrhage per slice. Patel et al. [40] modified 3D-CNN architecture for the automatic segmentation of ICH in non-contrast CT exams. This modified 3D CNN model was applied on a single subtype of hemorrhage for estimating the volume. Chang et al. [41] adopted the custom faster mask R-CNN algorithm to detect and segment hemorrhage. Although mask R-CNN produced excellent performance in segmentation with a high correlation score, the model estimated only the volume of IPH. Sharrock et al. [42] proposed the public source code for ICH segmentation, which is known as DeepBleed. It was trained for detecting the lesions of ICH and the occurrence of intraventricular hemorrhage (IVH) and SDH. In addition to the volume of lesion, its thickness is another key indicator used for surgical consideration. To the best of our knowledge, however, there is still no existing method mentioning both thickness and volume of various hemorrhage subtypes.
The method proposed in this paper differs from the aforementioned approaches. Herein, a framework is put forward to estimate both thickness and volume of hemorrhage subtypes through a quantitative assessment algorithm, with the output from two different deep neural networks used. The implementation of the method will be detailed in the next section.
3 Datasets and proposed method
3.1 Datasets
In this study, there are three datasets of brain hemorrhage used to train and evaluate the proposed method. Both public and private datasets are included, among which two datasets (RSNA 2019 Brain Hemorrhage Challenge and PhysioNet) are public datasets. The CMU-TBI is a private dataset. Each dataset is detailed as follows.
3.1.1 RSNA 2019 brain hemorrhage challenge dataset
The Radiological Society of North America (RSNA) [43] dataset can be found on Kaggle challenges. The objective of this competition is to identify the subtypes of ICH from brain CT scans. The dataset with annotations was collected and compiled by three research institutions located in the north and south of America. Due to the large amount of dataset comprised of over 25000 CT scans with five different subtypes of ICH labels, the competition attracted many developers and researchers from around the world to participate. The dataset involves six categories of brain hemorrhage including epidural hemorrhage (EDH), intraparenchymal hemorrhage (IPH), intraventricular hemorrhage (IVH), subarachnoid hemorrhage (SAH), subdural hemorrhage (SDH), and any existed hemorrhage. The raw data was stored in DICOM files. The DICOM format provides not only the pixel array of 512×512 but also header metadata. The total 755948 slices was divided into 740829 slices for the training set and 15119 slices for the test set, respectively.
3.1.2 PhysioNet
The PhysioNet [33] repository was collected from an Iraqi hospital during February and August 2018. There were two radiologists annotating the diagnosis of existing hemorrhage and ICH subtypes. The dataset is comprised of 82 CT scans including 46 male and 36 female scans with an average age of 27.8. There are approximate 34 slices included in each CT scan. A total of 2814 slices were extracted from these CT scans. The slices were split into 2233 slices for the training set and 581 slices for the test set.
3.1.3 CMU-TBI
This research has been granted ethical approval from the Ethics Committee of Faculty of Medicine, Chiang Mai University (CMU) and institutional review protocol. The head CT dataset includes the clinical data of 321 cases. There were about 30000 slices of a 1.5 mm thickness extracted from the Digital Imaging and Communications in Medicine (DICOM) series of CT Scans including 143 normal brains and 178 TBIs. The gender and age of patients are detailed in Table 1. The data of patients was collected from Maharaj Nakorn Chiang Mai Hospital. The slice numbers of detected EDH, SDH, IPH, SAH, and IVH were determined. The thickness of EDH and SDH, as well as the volume of IPH, were included as part of this dataset. Additionally, the data of those patients requiring surgical intervention was provided by the neuro-radiologists.
The 3D scan of a 1.5 mm slice thickness contains a set of 2D images ranging between 90 to 105 slices. Through our investigation, it was found out that hemorrhage appeared most commonly in the slices starting from slice number 20 to number 90. Therefore, the total number of slices was reduced to 19946 slices. Then, the dataset was split into 15956 for the training set and 3990 for the test set in this study.
The samples of each hemorrhage subtype in different three datasets used for training and testing the model are detailed as Table 2.
3.2 Proposed method
The objective of this study is to estimate the thickness and volume of hemorrhage. The thickness and volume of hemorrhage depend on the subtypes of hemorrhage, for example, the radiologists measure the thickness of EDH and SDH but the volume is determined through calculation for for the IPH type of hemorrhage. Herein, an optimal framework is proposed on the basis of a double-branch deep neural network and a quantitative assessment algorithm. With the fine-tuned multi-label classification performed and pre-trained multi-class segmentation algorithm adopted, the output features of both networks are treated as the input of the quantitative assessment algorithm to calculate the thickness and volume of different types of brain hemorrhage. The flowchart of our method is presented in Fig. 2. The raw CT Scans of the CMU-TBI database are in DICOM format including metadata and pixel data. The first branch refers to the process of training a multi-label classifier, while the second branch represents the task of multi-class segmentation. In order to achieve the final output of predicted thickness and volume, there are five major steps to go through, including data pre-processing and augmentation, multi-label classification, DICOM to NIfTI conversion, multi-class segmentation, and quantitative assessment. The details of each step will be presented in the following section.
3.2.1 Data pre-processing and augmentation
The multi-label classifier model was trained on the RSNA 2019 Brain CT Hemorrhage Challenge dataset before its integration into our method. The original pixel value of the images from the RSNA dataset is in the form of Hounsfield units (HU) representing the physical density of the tissue. HU allows radiologists to change the intensity windows during diagnosis. It consists of two separate windows: window center (WC) and window width (WW). According to the method suggested in [23], three HU windows are discussed depending on the exact type of issues. The window center (WC) and window width (WW) values of the three HU windows are expressed as:
-
Brain window (WC = 40, WW = 80)
-
Subdural window (WC = 80, WW = 200)
-
Soft tissue window (WC = 40, WW = 380)
The image of a HU window is grayscale. The results obtained from different HU windows were integrated into a single three-channel image as shown in Fig. 3. The original size of CT slices is 512× 512 pixels. The size of CT slices was changed into 256×256 pixels before data augmentation. As a result, the shape of the three-channel input for the classifier is 256×256×3.
With 25 percent of all images flipped horizontally and 10 percent of all images flipped vertically for data augmentation, the images captured from each side were cropped randomly between 0 to 25 pixels. Finally, the pixel values of all images were normalized into the range of [0,1].
3.2.2 Multi-label classifier
The up-to-date CNN architectures were refined for recognition, which were called EfficientNet [44] and EfficientNet with noisy student training [45]. The refined model was trained on the RSNA data and then taken as a pre-trained model. The original model architecture was modified by removing the last network layer and connecting it with the dropout layer (with dropout rate of 0.15). This is followed by a fully-connected layer with six output features that equal the number of categories in the RSNA dataset. The sigmoid activation function was applied after the fully connected layers. In this way, the final output provided the probability of ICH subtypes appearing in each image.
The EfficientNet models from B0 to B4 and EfficientNet with noisy student training models from B0 to B4 on the RSNA dataset with ten epochs were trained for comparison. The transfer learning mechanism was applied for this study through the following process. Firstly, EfficientNet-B2 was taken as a pre-trained network due to the highest accuracy (97%) and a reasonable number of parameters (7.77M). Secondly, the weight of the pre-trained model was transferred by fine-tuning the model with our CMU-TBI dataset. Finally, the multi-label classifier model provides the probability of each hemorrhage subtype that appears on each CT slice. The six output features represent the categories of hemorrhages including EDH, IPH, IVH, SAH, SDH, and an “exist or not” features. These features comprise one input for a quantitative assessment algorithm used to identify the types of hemorrhage. Multi-label log loss was taken as binary cross-entropy (BCE) loss for six output probabilities (the probabilities of five hemorrhagic subtypes and one probability of existed hemorrhage). The equation of BCE is expressed as:
where yt ∈{0,1} represents the ground truth label for class t, and \(\hat {y}_{t}\in \{0,1\}\) indicates the predicted class probability for a class t with the range of zero to one. By optimizing the BCE loss, the stochastic gradient descent technique with Adam [46] optimizer was applied at a 0.000125 learning rate. The batch sizes of 32 and 16 were adopted for the training set and test set, respectively. Each training session was processed for approximately two days on NVIDIA Tesla M10 GPU using Keras deep learning API. In this study, consideration was given only to the three subtypes (EDH, SDH, IPH) significant to decision-making on emergency surgery.
3.2.3 DICOM to NIfTI conversion
A multi-class segmentation method is required to assess the quantitative information of the hemorrhagic lesion. As one of the multi-class segmentation methods, DeepMedic [47] is based on a three-dimensional CNN architecture designed for the purpose of 3D segmentation. In the existing model, the format of NIfTI file is taken as the input data. Since NIfTI as a format of image is simpler than the DICOM format, it has been widely adopted for image processing and analysis [48]. Therefore, the conversion from DICOM to NIfTI is performed to prepare the data required for segmentation.
3.2.4 Multi-class segmentation
As a three-dimensional CNN for the accurate segmentation of brain lesion, DeepMedic [36] is comprised of eleven neural network layers. For the multi-class segmentation task on CT scans, an optimal DeepMedic model [37] was adopted. The model was modified to maintain the network architecture through residual blocks, batch normalization, and pre-activation blocks. Not only does the optimal DeepMedic outperform the existing medical image segmentation models such as U-Net [49] and UNet++ [50], it is also suitable for the tasks of multi-class segmentation. The optimal DeepMedic model was applied to obtain the multi-class segmentation mask for each slice of brain CT. Then, these output masks were used to classify the types and estimate the sizes of hemorrhage through the quantitative assessment algorithm. The samples of the predicted mask are shown in Fig. 4. The contours were separately colored according to each subtype of hemorrhage.
3.2.5 Quantitative assessment
Herein, a quantitative assessment algorithm is proposed. In the function, the output probabilities of fine-tuned EfficientNet-B2 are taken from branch #1 and the output mask of optimal DeepMedic is taken from branch #2 of a double-branch deep neural network, with every point on the output mask treated as the input. The network architectures of a double-branch deep neural network are shown in Fig. 5.
Network structure of a double-branch deep neural network based on fine-tuned EfficientNet-B2 and optimal DeepMedic. (a) Given the pre-processed 2D images and original 3D images, the feature extraction deep neural networks are covered by the gray area. (b-e) the details of each block in the main network
For each slice on a CT scan, the estimator is used to calculate the thickness and volume size of each contour separately. The contours are divided mainly into two groups. One is the contour that overlaps with the brain skull area including EDH and SDH types. The thickness estimator is applied to this group using Euclidean distance and the distance transform methods. The Euclidean distance provides the maximum and minimum lengths between a center point and other points. Based on these two lengths, the thickness ratio can be determined through calculation. The thickness ratio is a key factor to consider for distinguishing between EDH and SDH shapes. The distance transform is then applied to measure the thickness of a particular contour. The other is the contour of IPH inside the brain tissue area. The volume estimator function is applied to this group for the purpose of volume estimation.
The output of the quantitative assessment algorithm includes the thickness (in millimeter) and volume (in milliliter) estimated for of each subtype of hemorrhage. The pseudocode of the quantitative assessment algorithm applied for each CT scan is referred to Algorithm 1.

The probability threshold (PTEDH, PTSDH, PTIPH), thickness ratio (TR), and volume ratio (VR) are chosen based on accuracy and error calculation, respectively. The selection of probability threshold and thickness ratio is detailed in Section 5. The description of each function is explained as follows:
findSkullMask(s) - a function used to find the mask of the brain skull. With the input of each slice s, the function is expressed as Algorithm 2.

euclideanDistance(pz,pc) - a function that generates the Euclidean distance-vector D containing the distances between the center point pc and all of the other points in the contour c. The function is calculated by means of
distanceTransform(c) - a method used to replace each pixel p of the image with a distance to the nearest background pixel q. This method can be used to build the distance map DM. The output value of distance transformation is approximately half the actual lesion width. The distance map DM is expressed as
where I(q) represents the pixel value of q.
findVolume(w,h,ps) - a function intended to estimate the volume of hemorrhagic lesions. This function is derived from the ABC/2 method [51]. The ABC/2 is a technique proposed by Kothari et al. to calculate the volume of hemorrhage, where A represents the value of maximum length (in cm), B indicates the width perpendicular to A on the same head CT slice, and C denotes the number of slices multiplied by the thickness of slice. Thus, the findVolume(w,h,ps) function is expressed as (4).
In this study, slice thickness is set to 1.5 mm.
4 Evaluation
The performance of the proposed method and that of the baseline methods are compared. Then, comparison results are categorized mainly into three sets of evaluations. In the first one, the classification results are compared between our fine-tuned EfficientNet-B2 and baseline methods. In the second one, the performance of the optimal DeepMedic is discussed using segmentation metrics. The last one demonstrates the results of classification and estimation for the size of hemorrhagic lesions based on a double-branch deep neural network built on a private CMU-TBI dataset.
4.1 The performance evaluations between fine-tuned EfficientNet-B2 and baseline methods
There are five metrics used to evaluate the classification performance including precision, sensitivity, specificity, f1-score and accuracy. Each of them can be calculated using the following equations:
Precision
Sensitivity or recall
Specificity
F1-score
Accuracy
where TP represents a true positive value, TN refers to a true negative value, FP denotes a false positive value, and FN indicates a false negative value. All metrics are converted into the percentage unit.
There are various baseline methods, for example, decision tree-based Projection Profile [17], ICH UNet [33], and UNet++ [50]. According to Table 3, the fine-tuned EfficientNet-B2 outperforms the baseline methods on the CMU-TBI dataset in terms of classification. The models were trained for 100 epochs to obtain the results. The scores of the fine-tuned EfficientNet-B2 are mostly higher compared to ICH U-Net and UNet++ methods. The specificity rate of UNet++ is the lowest due to large proportion of false-positive diagnosis. In other words, the model predicts normal brain wrongly as hemorrhagic lesion.
Figure 6 shows the accuracy and loss charts of fine-tuned EfficientNet-B2 on the CMU-TBI dataset. During the training process, the model achieves higher accuracy and lower loss than in the testing process. Through comparison with the performance during the training process, it can be discovered that the accuracy and loss during the testing process converge and maintain consistency after 40 epochs. The output suggests that the performance of the model during the testing process did not improve with the increase in epoch.
PhysioNet is the public dataset that used in this study to evaluate the performance of our model. Even though the sensitivity score of our model is lower relative to U-Net and UNet++, the other metrics surpass baseline methods as shown in Table 4.
4.2 The performance evaluation between optimal DeepMedic algorithm and baseline methods
In order to quantify the performance of models in segmentation, there are two methods are adopted, including Jaccard Index which is also known as Intersection over Union (IoU) and Dice score (Dice similarity coefficient). Jaccard Index is an effective metric intended to measure the accuracy between the predicted output mask and ground truth mask by computing the overlapping area of those masks under the union area of both masks. With Y representing ground truth segmentation and \(\hat {Y}\) referring to the predicted output of methods, the Jaccard Index and Dice score are written as (10) and (11), respectively.
Where ∩ denotes intersection and ∪ represents the union of two segmentations Y and \(\hat {Y}\), while |⋅| indicates the summation result of the argument. The values of Y and \(\hat {Y}\) range from 0 to 1.
The ICH UNet [33], DeepBleed [42], and UNet++ [50] were treated as the baseline methods. The Jaccard Index and Dice Score of baseline methods and our method on the publicly accessible PhysioNet dataset were calculated, while the metrics were calculated on the test set including 581 slices. Consistent with the experimental evaluation shown in Table 5, the optimal DeepMedic outperforms most of the baseline methods in both Jaccard Index and Dice Score except UNet++. However, the UNet++ is incapable of multi-class segmentation, with low sensitivity score achieved.
Figure 7 shows the segmentation regions and evaluation metrics of the baseline model (UNet++) and our method on the PhysioNet dataset. In the first row of region output, the red line represents the supervised mask as provided with the dataset and the green line refers to the predicted mask from the UNet++ model. The regions of the supervised mask and predicted mask are largely overlapped. However, there are some false-positive regions detected. The results in the second row are from ground truth and our method. The predicted regions of our approach exclude many false-positive regions, which however differ from the baseline model. Moreover, the optimal DeepMedic model and baseline model were tested on the CMU-TBI dataset as shown in Fig. 8. According to the output of segmentation, our method covers more types of hemorrhagic lesion including the small region (last image) than the UNet++ model.
4.3 The performance evaluation of our double-branch deep learning network with quantitative assessment algorithm on each subtype of hemorrhage in CMU-TBI dataset
In this part, a comparison was performed between the results obtained from the classification of types of hemorrhagic lesion. Additionally, the error metrics of thickness and volume calculation were discussed. The experimental analysis was conducted through the classification task by observing the classification metrics of our method on the CMU-TBI dataset. The 56 CT scans with obviously differentiated lesion types from a total of 178 scans were treated as a validation set. The validation set is comprised of 3130 slices. Each slice shows only one type of hemorrhage, that is, either EDH, SDH, or IPH. Table 6 shows the comparison of our method on each type of hemorrhage in different measurements. The hybrid method based on quantitative assessment algorithm achieves the highest accuracy of 96.54 percent when SDH is classified. The average accuracy is 96.21 percent for the three types of hemorrhage.
4.4 The thickness and volume difference of EDH, SDH, and IPH between true and predicted values from our method
A total of 56 CT scans in the validation set obtained from the CMU-TBI dataset were included to calculate the difference in thickness and volume between the true values provided by the doctor and the values estimated by using our method. Figure 9 shows the Bland-Altman plots of agreement between ground truth and predicted values. The mean difference of thickness is 2.99 mm (-0.42 to 6.42) for EDH and 0.97 mm (-2.41 to 4.35) for SDH. The mean different volume of IPH is 0.43 mL (-4.74 to 5.61).
5 Ablation study
5.1 Probability threshold selection
The probability threshold is a parameter required to select the model that achieves the highest accuracy. The probabilities ranging between 0.1 to 0.9 for each subtype of hemorrhage are evaluated, as shown in Fig. 10. The best probability threshold for the subtypes of EDH (PTEDH), SDH (PTSDH), and IPH (PTIPH) is 0.5, 0.2, and 0.1, respectively.
5.2 Thickness and volume ratio selection
The optimal thickness ratio (TR) for EDH and SDH subtypes and volume ratio (VR) for IPH subtype can be identified by the minimum Mean Absolute Error (MAE) while the model is tested using different ratios. The ratios selected for testing the model range from 1 to 25. The MAE is expressed as (12).
q represents the ground truth quantitative information (thickness or volume) provided by experts, \(\hat {q}_{i}\) indicates predicted quantitative information, and n denotes the number of lesions in each subtype.
In order to find the optimal TR and VR, the true-positive MAE (MAETP) and false-positive MAE (MAEFP) of each subtype are obtained. The MAETP evaluates the error between ground truth and predicted quantitative information within the same category, while the MAEFP is used to calculate the error between ground truth and the predicted quantitative information of different types. The optimal TR is defined as the ratio that provides a minimum average MAE of thickness values in EDH and SDH subtypes as calculated using the following equation.
MAEAVG(EDH,SDH) represents the average MAE of thickness values in EDH and SDH subtypes defined as
MAETP(EDH) is true-positive MAE for EDH, MAEFP (EDH) is false-positive MAE for EDH, MAETP(SDH) is true-positive MAE for SDH, and MAEFP(SDH) is false-positive MAE for SDH. Figure 11 shows MAETP and MAEFP for different thickness ratios of our method and the original DeepMedic.
As shown in Fig. 11a, the true-positive MAE of our method is clearly comparable to the traditional DeepMedic method. In spite of this, our method can also achieve less false-positive and overall MAE than the baseline approach as shown in Fig. 11b and c, respectively. The TR was set to 20 as the lowest point in average MAE.
The optimal VR is referred to as the ratio that provides a minimum average MAE of volume values in the subtype of IPH. It can be expressed as the following equation.
MAEAVG(IPH) refers to the average MAE of volume values in IPH subtype which is defined as
MAETP(IPH) is true-positive MAE for IPH, and MAEFP(IPH) is false-positive MAE for IPH. Figure 12 shows the MAETP and MAEFP for different volume ratios of our method and the original DeepMedic.
The method proposed in this study improved the true-positive MAE when the ratio value increased, as shown in Fig. 12a. The MAE of volume measurement can als be reduced significantly, as shown in Fig. 12b. The VR was set to 24 as the lowest point in average MAE.
6 Conclusion
The prior studies on automated assessment of head CT images after TBI are limited to the undifferentiated detection of hemorrhage different lesions, with no quantitative assessment conducted for the volumetric analysis. For this reason, the accurate detection and quantification of lesion volumes are essential for improving the understanding of those influencing factors in lesion progression and targeted medical treatment. In this study, an optimal deep learning framework is proposed, which can not only identify the subtypes of hemorrhages but also assist the clinically relevant quantitative assessment of thickness and volume. The proposed method is integrated with a fine-tuned multi-label classifier (EfficientNet-B2), an optimal multi-class segmentation model (DeepMedic), and our quantitative assessment algorithm. The fine-tuned EfficientNet-B2 model can achieve the highest accuracy with 98.62 percent on the CMU-TBI dataset in comparison with two baseline models, namely, ICH U-Net and UNet++.
In addition, the Jaccard Index and Dice score of our method are calculated using the output from the optimal DeepMedic. The model shows a comparable Jaccard Index and Dice score to the baseline methods on a PhysioNet dataset.
The quantitative assessment algorithm takes the probabilities of each hemorrhage subtype from a fine-tuned multi-label classifier and hemorrhage contours from the optimal multi-class segmentation model as inputs. In order to differentiate hemorrhagic lesions, our method is also assessed for each subtype of hemorrhage. The model is tested to classify EDH, SDH, and IPH separately on a validation set of the CMU-TBI dataset. According to the test results, our method performs best in classification for SDH type with a 96.54 percent accuracy. The average accuracy is 96.21 percent for the three subtypes of hemorrhage.
The thickness and volume of hemorrhagic lesions are computed by means of distance transform and the commonly applied volume evaluation ABC/2 functions. The differences between ground truth and predicted lesions (of thickness and volume) are indicated by Bland-Altman plots. The predicted EDH, SDH thickness and IPH volume overestimated the true values by 2.99 mm, 0.97 mm, and 0.43 mL, sequentially. Moreover, our method can reduce the false-positive mean absolute error of both thickness and volume assessments more significantly than the traditional DeepMedic multi-class segmentation approach.
With this fully automated method applied, the process of decision-making on surgery can be accelerated and the shortage of radiologists can be addressed for rural medical institutions. In the future, it is necessary to improve the technique based on the aforementioned surgical consideration. Integrating the research into clinical practice requires various additional functions such as skull fracture detection and midline shift measurement. For the better understanding and prognostication of lesions, it is essential to conduct adequate validation on other subtypes of hemorrhage.
References
Savard J, Ouellet MC (2019) Handbook of sleep disorders in medical conditions. Academic Press, New York
Mansour RF, Aljehane NO (2021) An optimal segmentation with deep learning based inception network model for intracranial hemorrhage diagnosis. Neural Comput Appl :1–13
Jullabussapa N, Khwanngern K, Pateekhum C, Angkurawaranon C, Angkurawaranon S (2020) Ct-based measurements of facial parameters of healthy children and adolescents in thailand. Am J Neuroradiol 41(10):1937–1942
Gupta V, Prabhakar A, Yadav M, Khandelwal N (2019) Computed tomography imaging-based normative orbital measurement in indian population. Indian J Ophthalmol 67(5):659
Sirimaharaj W, Kanpuan W, Angkurawaranon S (2019) The relationship between external bony defects and widened lateral interorbital distance in frontoethmoidal encephalomeningocele. J Cranio-Maxillofac Surg 47(10):1563–1568
Liu Y, Wang H, Gu Y, Lv X (2019) Image classification toward lung cancer recognition by learning deep quality model. J Vis Commun Image Represent 63:102570
Makaju S, Prasad P, Alsadoon A, Singh A, Elchouemi A (2018) Lung cancer detection using ct scan images. Procedia Comput Sci 125:107–114
Ai Z (2019) Quantitative ct study of martial arts sports injuries based on image quality. J Vis Commun Image Represent 60:417–425
Huang Z, Liu X, Wang R, Zhang M, Zeng X, Liu J, Yang Y, Liu X, Zheng H, Liang D et al (2020) Fanet: fast assessment network for the novel coronavirus (covid-19) pneumonia based on 3d ct imaging and clinical symptoms. Appl Intell :1–12
Gao K, Su J, Jiang Z, Zeng LL, Feng Z, Shen H, Rong P, Xu X, Qin J, Yang Y et al (2021) Dual-branch combination network (dcn): Towards accurate diagnosis and lesion segmentation of covid-19 using ct images. Med Image Anal 67:101836
Ahuja S, Panigrahi BK, Dey N, Rajinikanth V, Gandhi TK (2021) Deep transfer learning-based automated detection of covid-19 from lung ct scan slices. Appl Intell 51(1):571–585
Bazarian JJ, Biberthaler P, Welch RD, Lewis LM, Barzo P, Bogner-Flatz V, Brolinson PG, Büki A, Chen JY, Christenson RH et al (2018) Serum gfap and uch-l1 for prediction of absence of intracranial injuries on head ct (alert-tbi): a multicentre observational study. Lancet Neurol 17(9):782–789
Rosen CB, Luy DD, Deane MR, Scalea TM, Stein DM (2018) Routine repeat head ct may not be necessary for patients with mild tbi. Trauma Surg Acute Care Open 3(1):e000129
Honda M, Ichibayashi R, Yokomuro H, Yoshihara K, Masuda H, Haga D, Seiki Y, Kudoh C, Kishi T (2016) Early cerebral circulation disturbance in patients suffering from severe traumatic brain injury (tbi): a xenon ct and perfusion ct study. Neurol Med Chir 56(8):501–509
Fong R, Konakondla S, Schirmer C, Lacroix M (2017) Surgical interventions for severe traumatic brain injury. J Emerg Crit Care Med 1(10):28–28
Bullock MR, Chesnut R, Ghajar J, Gordon D, Hartl R, Newell DW, Servadei F, Walters BC, Wilberger JE (2006) Introduction. Neurosurgery 58
Thay S, Aimmanee P, Uyyanavara B, Rukskul P (2018) Fast hemorrhage detection in brain ct scan slices using projection profile based decision tree. In: Proceedings of the 2018 international conference on intelligent information technology, pp 18–21
Srivastava DK, Sharma B, Singh A (2018) Classification of hematomas in brain ct images using support vector machine. In: Information and communication technology for sustainable development. Springer, pp 375–385
Liu J, Xu H, Chen Q, Zhang T, Sheng W, Huang Q, Song J, Huang D, Lan L, Li Y et al (2019) Prediction of hematoma expansion in spontaneous intracerebral hemorrhage using support vector machine. EBioMedicine 43:454–459
Chilamkurthy S, Ghosh R, Tanamala S, Biviji M, Campeau NG, Venugopal VK, Mahajan V, Rao P, Warier P (2018) Deep learning algorithms for detection of critical findings in head ct scans: a retrospective study. Lancet 392(10162):2388–2396
Jnawali K, Arbabshirani MR, Rao N, Patel AA (2018) Deep 3d convolution neural network for ct brain hemorrhage classification. In: Medical imaging 2018: Computer-aided diagnosis, international society for optics and photonics, vol 10575, p 105751C
Lee H, Yune S, Mansouri M, Kim M, Tajmir SH, Guerrier CE, Ebert SA, Pomerantz SR, Romero JM, Kamalian S et al (2019) An explainable deep-learning algorithm for the detection of acute intracranial haemorrhage from small datasets. Nat Biomed Eng 3(3):173–182
Burduja M, Ionescu RT, Verga N (2020) Accurate and efficient intracranial hemorrhage detection and subtype classification in 3d ct scans with convolutional and long short-term memory neural networks. Sensors 20(19):5611
Ko H, Chung H, Lee H, Lee J (2020) Feasible study on intracranial hemorrhage detection and classification using a cnn-lstm network. In: 2020 42nd annual international conference of the IEEE engineering in medicine & biology society (EMBC). IEEE, pp 1290–1293
Nguyen NT, Tran DQ, Nguyen NT, Nguyen HQ (2020) A CNN-LSTM architecture for detection of intracranial hemorrhage on CT scans. arXiv:2005.10992
He J (2020) Automated detection of intracranial hemorrhage on head computed tomography with deep learning. In: Proceedings of the 2020 10th international conference on biomedical engineering and technology, pp 117–121
Wang H, Peng J, Zhao Y, Fu X (2020a) Multi-path deep cnns for fine-grained car recognition. IEEE Trans Veh Technol 69(10):10484–10493
Wang H, Peng J, Chen D, Jiang G, Zhao T, Fu X (2020b) Attribute-guided feature learning network for vehicle reidentification. IEEE Multimed 27(4):112–121
Zhang T, Sun X, Li X, Yi Z (2021) Image generation and constrained two-stage feature fusion for person re-identification. Appl Intell. https://doi.org/10.1007/s10489-021-02271-z
Phaphuangwittayakul A, Guo Y, Ying F (2021) Fast adaptive meta-learning for few-shot image generation. IEEE Trans Multimed. https://doi.org/10.1109/TMM.2021.3077729
Farzaneh N, Soroushmehr SR, Williamson CA, Jiang C, Srinivasan A, Bapuraj JR, Ward KR, Korley FK, Najarian K (2017) Automated subdural hematoma segmentation for traumatic brain injured (tbi) patients. In: 2017 39th annual international conference of the IEEE engineering in medicine and biology society (EMBC). IEEE, pp 3069–3072
Remedios SW, Roy S, Bermudez C, Patel MB, Butman JA, Landman BA, Pham DL (2020) Distributed deep learning across multisite datasets for generalized ct hemorrhage segmentation. Med Phys 47(1):89–98
Hssayeni MD, Croock MS, Salman AD, Al-khafaji HF, Yahya ZA, Ghoraani B (2020) Intracranial hemorrhage segmentation using a deep convolutional model. Data 5(1):14
Kuo W, Hane C, Mukherjee P, Malik J, Yuh EL (2019) Expert-level detection of acute intracranial hemorrhage on head computed tomography using deep learning. Proc Natl Acad Sci 116(45):22737–22745
Monteiro M, Kamnitsas K, Ferrante E, Mathieu F, McDonagh S, Cook S, Stevenson S, Das T, Khetani A, Newman T, et al. (2019) Tbi lesion segmentation in head ct: Impact of preprocessing and data augmentation. In: International MICCAI brainlesion workshop. Springer, pp 13–22
Kamnitsas K, Ledig C, Newcombe VF, Simpson JP, Kane AD, Menon DK, Rueckert D, Glocker B (2017) Efficient multi-scale 3d cnn with fully connected crf for accurate brain lesion segmentation. Med Image Anal 36:61–78
Monteiro M, Newcombe VF, Mathieu F, Adatia K, Kamnitsas K, Ferrante E, Das T, Whitehouse D, Rueckert D, Menon DK et al (2020) Multiclass semantic segmentation and quantification of traumatic brain injury lesions on head ct using deep learning: an algorithm development and multicentre validation study. Lancet Digit Health 2(6):e314–e322
Chi Fl, Lang Tc, Sun Sj, Tang Xj, Xu SY, Zheng Hb, Zhao HS (2014) Relationship between different surgical methods, hemorrhage position, hemorrhage volume, surgical timing, and treatment outcome of hypertensive intracerebral hemorrhage. World J Emerg Med 5(3):203
Jain S, Vyvere TV, Terzopoulos V, Sima DM, Roura E, Maas A, Wilms G, Verheyden J (2019) Automatic quantification of computed tomography features in acute traumatic brain injury. J Neurotrauma 36(11):1794–1803
Patel A, Schreuder FH, Klijn CJ, Prokop M, van Ginneken B, Marquering HA, Roos YB, Baharoglu MI, Meijer FJ, Manniesing R (2019) Intracerebral haemorrhage segmentation in non-contrast ct. Sci Rep 9(1):1–11
Chang PD, Kuoy E, Grinband J, Weinberg BD, Thompson M, Homo R, Chen J, Abcede H, Shafie M, Sugrue L et al (2018) Hybrid 3d/2d convolutional neural network for hemorrhage evaluation on head ct. Am J Neuroradiol 39(9):1609–1616
Sharrock MF, Mould WA, Ali H, Hildreth M, Awad IA, Hanley DF, Muschelli J (2020) 3d deep neural network segmentation of intracerebral hemorrhage: Development and validation for clinical trials. Neuroinformatics :1–13
Flanders AE, Prevedello LM, Shih G, Halabi SS, Kalpathy-Cramer J, Ball R, Mongan JT, Stein A, Kitamura FC, Lungren MP et al (2020) Construction of a machine learning dataset through collaboration: the rsna 2019 brain ct hemorrhage challenge. Radiol Artif Intell 2(3):e190211
Tan M, Le Q (2019) Efficientnet: Rethinking model scaling for convolutional neural networks. In: International conference on machine learning, PMLR, pp 6105–6114
Xie Q, Luong MT, Hovy E, Le QV (2020) Self-training with noisy student improves imagenet classification. In: Proceedings of the IEEE/CVF conference on computer vision and pattern recognition, pp 10687–10698
Kingma DP, Ba J (2015) Adam: A method for stochastic optimization. In: 3rd International conference on learning representations, ICLR
Kamnitsas K, Ferrante E, Parisot S, Ledig C, Nori AV, Criminisi A, Rueckert D, Glocker B (2016) Deepmedic for brain tumor segmentation. In: International workshop on brainlesion: Glioma, multiple sclerosis, stroke and traumatic brain injuries. Springer, pp 138–149
Li X, Morgan PS, Ashburner J, Smith J, Rorden C (2016) The first step for neuroimaging data analysis: Dicom to nifti conversion. J Neurosci Methods 264:47–56
Ronneberger O, Fischer P, Brox T (2015) U-net: Convolutional networks for biomedical image segmentation. In: International conference on medical image computing and computer-assisted intervention. Springer, pp 234–241
Zhou Z, Siddiquee MMR, Tajbakhsh N, Liang J (2018) Unet++: A nested u-net architecture for medical image segmentation. In: Deep learning in medical image analysis and multimodal learning for clinical decision support. Springer, pp 3–11
Kothari RU, Brott T, Broderick JP, Barsan WG, Sauerbeck LR, Zuccarello M, Khoury J (1996) The abcs of measuring intracerebral hemorrhage volumes. Stroke 27(8):1304–1305
Acknowledgements
This research is financially supported by The National Key Research and Development Program of China (grant number 2018YFC0807105) and Science and Technology Committee of Shanghai Municipality (STCSM) (grant numbers 17DZ1101003, 18511106602 and 18DZ2252300). Partially Supported by Open Funding Project of the State Key Laboratory of Bioreactor Engineering, East China University of Science and Technology, Shanghai, China. Study resources and dataset were supported by International College of Digital Innovation (ICDI) and Faculty of Medicine (grant number 143-2562), Chiang Mai University.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of Interests
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Phaphuangwittayakul, A., Guo, Y., Ying, F. et al. An optimal deep learning framework for multi-type hemorrhagic lesions detection and quantification in head CT images for traumatic brain injury. Appl Intell 52, 7320–7338 (2022). https://doi.org/10.1007/s10489-021-02782-9
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10489-021-02782-9