Abstract
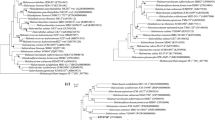
The current species of Halosegnis and Salella within the class Halobacteria are closely related based on phylogenetic, phylogenomic, and comparative genomic analyses. The Halosegnis species showed 99.8–100.0% 16S rRNA and 96.6–99.6% rpoB′ gene similarities to the Salella species, respectively. Phylogenetic and phylogenomic analyses showed that Salella cibi CBA1133T, the sole species of Salella, formed a single tight cluster with Halosegnis longus F12-1T, then with Halosegnis rubeus F17-44T. The average nucleotide identity (ANI), digital DNA–DNA hybridization (dDDH), and average amino acid identity (AAI) values between Salella cibi CBA1133T and Halosegnis longus F12-1T were 99.2, 94.2, and 98.6%, respectively, much higher than the thresholds for species demarcation. This genome-based classification revealed that the genus Salella should be merged with Halosegnis, and Salella cibi should be a later heterotypic synonym of Halosegnis longus. Halophilic archaeal strains DT72T, DT80T, DT85T, and DT116T, isolated from the saline soil of a tidal flat in China, were subjected to polyphasic taxonomic characterization. The phenotypic, chemotaxonomic, phylogenetic, and phylogenomic features indicated that strains DT72T (= CGMCC 1.18925T = JCM 35418T), DT80T (= CGMCC 1.18926T = JCM 35419T), DT85T (= CGMCC 1.19049T = JCM 35605T), and DT116T (= CGMCC 1.19045T = JCM 35606T) represent four novel species of the genera Halorussus, Halosegnis and Haloglomus, respectively, for which the names, Halorussus caseinilyticus sp. nov., Halorussus lipolyticus sp. nov., Halosegnis marinus sp. nov., and Haloglomus litoreum sp. nov., are proposed.


Similar content being viewed by others
Data availability
The sequences determined in this study were deposited and available in the NCBI GenBank.
References
Aziz RK, Bartels D, Best AA, DeJongh M, Disz T, Edwards RA, Formsma K, Gerdes S, Glass EM, Kubal M, Meyer F, Olsen GJ, Olson R, Osterman AL, Overbeek RA, McNeil LK, Paarmann D, Paczian T, Parrello B, Pusch GD, Reich C, Stevens R, Vassieva O, Vonstein V, Wilke A, Zagnitko O (2008) The RAST server: rapid annotations using subsystems technology. BMC Genom 9:75. https://doi.org/10.1186/1471-2164-9-75
Bao CX, Li SY, Xin YJ, Hou J, Cui HL (2022) Natrinema halophilum sp. nov., Natrinema salinisoli sp nov, Natrinema amylolyticum sp. nov. and Haloterrigena alkaliphila sp. nov. four extremely halophilic archaea isolated from salt mine saline soil and salt lake. Int J Syst Evol Microbiol 72:005385. https://doi.org/10.1099/ijsem.0.005385
Chun J, Oren A, Ventosa A, Christensen H, Arahal DR, da Costa MS, Rooney AP, Yi H, Xu XW, De Meyer S, Trujillo ME (2018) Proposed minimal standards for the use of genome data for the taxonomy of prokaryotes. Int J Syst Evol Microbiol 68(1):461–466. https://doi.org/10.1099/ijsem.0.002516
Cui HL, Dyall-Smith ML (2021) Cultivation of halophilic archaea (class Halobacteria) from thalassohaline and athalassohaline environments. Mar Life Sci Technol 3(2):243–251. https://doi.org/10.1007/s42995-020-00087-3
Cui HL, Zhou PJ, Oren A, Liu SJ (2009) Intraspecific polymorphism of 16S rRNA genes in two halophilic archaeal genera, Haloarcula and Halomicrobium. Extremophiles 13(1):31–37. https://doi.org/10.1007/s00792-008-0194-2
Cui HL, Gao X, Yang X, Xu XW (2010) Halorussus rarus gen. nov., sp. nov., a new member of the family Halobacteriaceae isolated from a marine solar saltern. Extremophiles 14(6):493–499. https://doi.org/10.1007/s00792-010-0329-0
Cui C, Han D, Hou J, Cui HL (2023) Genome-based classification of the class Halobacteria and description of Haladaptataceae fam. nov. and Halorubellaceae fam. nov. Int J Syst Evol Microbiol 73:005984. https://doi.org/10.1099/ijsem.0.005984
Deshmukh UB, Oren A (2023) Proposal of Eoetvoesiella gen. nov., Paludihabitans gen. nov., Rivihabitans gen. nov. and Salella gen. nov. as replacement names for the illegitimate prokaryotic generic names Eoetvoesia, Paludicola, Rivicola and Sala, respectively. Int J Syst Evol Microbiol 73(5):005901. https://doi.org/10.1099/ijsem.0.005901
Durán-Viseras A, Sánchez-Porro C, Ventosa A (2020) Haloglomus irregulare gen. nov., sp. nov., a New Halophilic Archaeon isolated from a marine saltern. Microorganisms 8(2):206. https://doi.org/10.3390/microorganisms8020206
Durán-Viseras A, Andrei AŞ, Vera-Gargallo B, Ghai R, Sánchez-Porro C, Ventosa A (2021a) Culturomics-based genomics sheds light on the ecology of the new haloarchaeal genus Halosegnis. Environ Microbiol 23(7):3418–3434. https://doi.org/10.1111/1462-2920.15082
Durán-Viseras A, Sánchez-Porro C, Ventosa A (2021b) Genomic insights into new species of the genus halomicroarcula reveals potential for new osmoadaptative strategies in halophilic archaea. Front Mar Sci 12:751746. https://doi.org/10.3389/fmicb.2021.751746/full
Dussault HP (1955) An improved technique for staining red halophilic bacteria. J Bacteriol 70:484–485. https://doi.org/10.1128/jb.70.4.484-485.1955
Felsenstein J (1981) Evolutionary trees from DNA sequences: a maximum likelihood approach. J Mol Evol 17:368–376. https://doi.org/10.1007/BF01734359
Göker M, Oren A (2023) Valid publication of four additional phylum names. Int J Syst Evol Microbiol 73(9):006024. https://doi.org/10.1099/ijsem.0.006024
Hu Y, Ma X, Li XX, Tan S, Cheng M, Hou J, Cui HL (2023) Natrinema caseinilyticum sp. nov., Natrinema gelatinilyticum sp. nov., Natrinema marinum sp. nov., Natrinema zhouii sp. nov., extremely halophilic archaea isolated from marine environments and a salt mine. Extremophiles 27(1):9. https://doi.org/10.1007/s00792-023-01294-3
Kanehisa M, Goto S, Hattori M, Aoki-Kinoshita KF, Itoh M, Kawashima S, Katayama T, Araki M, Hirakawa M (2006) From genomics to chemical genomics: new developments in KEGG. Nucleic Acids Res 34:D354–D357. https://doi.org/10.1093/nar/gkj102
Kilic V, Kilic GA, Kutlu HM, Martínez-Espinosa RM (2017) Nitrate reduction in Haloferax alexandrinus: the case of assimilatory nitrate reductase. Extremophiles 21(3):551–561. https://doi.org/10.1007/s00792-017-0924-4
Kim BS, Kim BK, Lee JH, Kim M, Lim YW, Chun J (2008) Rapid phylogenetic dissection of prokaryotic community structure in tidal flat using pyrosequencing. J Microbiol 46:357–363. https://doi.org/10.1007/s12275-008-0071-9
Kim M, Oh HS, Park SC, Chun J (2014) Towards a taxonomic coherence between average nucleotide identity and 16S rRNA gene sequence similarity for species demarcation of prokaryotes. Int J Syst Evol Microbiol 64:346–351. https://doi.org/10.1099/ijs.0.059774-0
Li SY, Xin YJ, Bao CX, Hou J, Cui HL (2021) Haloprofundus salilacus sp. nov., Haloprofundus halobius sp. nov. and Haloprofundus salinisoli sp. nov.: three extremely halophilic archaea isolated from salt lake and saline soil. Extremophiles 26(1):6. https://doi.org/10.1007/s00792-021-01255-8
Luo C, Rodriguez-R LM, Konstantinidis KT (2014) MyTaxa: an advanced taxonomic classifier for genomic and metagenomic sequences. Nucleic Acids Res 42:e73. https://doi.org/10.1093/nar/gku169
Meier-Kolthoff JP, Auch AF, Klenk HP, Göker M (2013) Genome sequence-based species delimitation with confidence intervals and improved distance functions. BMC Bioinformatics 14:60. https://doi.org/10.1186/1471-2105-14-60
Nwankwo C, Hou J, Cui H-L (2023) Extracellular proteases from halophiles: diversity and application challenges. Appl Microbiol Biotechnol 107(19):5923–5934. https://doi.org/10.1007/s00253-023-12721-y
Oren A, Ventosa A, Grant WD (1997) Proposed minimal standards for description of new taxa in the order Halobacteriales. Int J Syst Evol Microbiol 47:233–238. https://doi.org/10.1099/00207713-47-1-233
Parks DH, Chuvochina M, Waite DW, Rinke C, Skarshewski A, Chaumeil PA, Hugenholtz P (2018) A standardized bacterial taxonomy based on genome phylogeny substantially revises the tree of life. Nat Biotechnol 36:996–1004. https://doi.org/10.1038/nbt.4229
Parte AC, Sardà Carbasse J, Meier-Kolthoff JP, Reimer LC, Göker M (2020) List of prokaryotic names with standing in nomenclature (LPSN) moves to the DSMZ. Int J Syst Evol Microbiol 70:5607–5612. https://doi.org/10.1099/ijsem.0.004332
Richter M, Rosselló-Móra R, Oliver Glöckner F, Peplies J (2016) JSpeciesWS: a web server for prokaryotic species circumscription based on pairwise genome comparison. Bioinformatics 32:929–931. https://doi.org/10.1093/bioinformatics/btv681
Sakaguchi M (2020) Diverse and common features of trehalases and their contributions to microbial trehalose metabolism. Appl Microbiol Biotechnol 104(5):1837–1847. https://doi.org/10.1007/s00253-019-10339-7
Serrano S, Mendo S, Caetano T (2022) Haloarchaea have a high genomic diversity for the biosynthesis of carotenoids of biotechnological interest. Res Microbiol 173(3):103919. https://doi.org/10.1016/j.resmic.2021.103919
Shrestha P, Karmacharya J, Han S-R, Lee JH, Park H, Oh T-J (2022) Complete Genome sequence and comparative genome analysis of Variovorax sp. strains PAMC28711, PAMC26660, and PAMC28562 and trehalose metabolic pathways in antarctica isolates. Int J Microbiol 2022:5067074. https://doi.org/10.1155/2022/5067074
Song HS, Kim J, Kim YB, Lee SH, Whon TW, Roh SW (2022) Sala cibi gen. nov., sp. nov., an extremely halophilic archaeon isolated from solar salt. J Microbiol 60(9):899–904. https://doi.org/10.1007/s12275-022-2137-5
Sun YP, Wang BB, Zheng XW, Wu ZP, Hou J, Cui HL (2022) Description of Halosolutus amylolyticus gen. nov., sp. nov., Halosolutus halophilus sp. nov. and Halosolutus gelatinilyticus sp. nov., and genome-based taxonomy of genera Natribaculum and Halovarius. Int J Syst Evol Microbiol 72(10):005598. https://doi.org/10.1099/ijsem.0.005598
Sun YP, Wang BB, Wu ZP, Zheng XW, Hou J, Cui HL (2023) Halorarius litoreus gen. nov., sp. nov., Halorarius halobius sp. nov., Haloglomus halophilum sp. nov., Haloglomus salinum sp. nov., and Natronomonas marina sp. nov., extremely halophilic archaea isolated from tidal flat and marine solar salt. Front Mar Sci 10:1105929. https://doi.org/10.3389/fmars.2023.1105929
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30:2725–2729. https://doi.org/10.1093/molbev/mst197
Tan S, Cheng M, Li XX, Hu Y, Ma X, Hou J, Cui HL (2023) Natronosalvus halobius gen. nov., sp. nov., Natronosalvus caseinilyticus sp. nov., Natronosalvus vescus sp. nov., Natronosalvus rutilus sp. nov. and Natronosalvus amylolyticus sp. nov., halophilic archaea isolated from salt lakes and soda lakes. Int J Syst Evol Microbiol 73(9):006036. https://doi.org/10.1099/ijsem.0.006036
Tatusova T, DiCuccio M, Badretdin A, Chetvernin V, Nawrocki EP, Zaslavsky L, Lomsadze A, Pruitt KD, Borodovsky M, Ostell J (2016) NCBI prokaryotic genome annotation pipeline. Nucleic Acids Res 44(14):6614–6624. https://doi.org/10.1093/nar/gkw569
Xin YJ, Bao CX, Li SY, Hu XY, Zhu L, Wei W, Hou J, Cui HL (2022) Genome-based taxonomy of genera Halomicrobium and Halosiccatus, and description of Halomicrobium salinisoli sp. nov. Syst Appl Microbiol 45(3):126308. https://doi.org/10.1016/j.syapm.2022.126308
Xu L, Dong Z, Fang L, Luo Y, Wei Z, Guo H, Zhang G, Gu YQ, Coleman-Derr D, Xia Q, Wang Y (2019) OrthoVenn2: a web server for whole-genome comparison and annotation of orthologous clusters across multiple species. Nucleic Acids Res 47(W1):W52–W58. https://doi.org/10.1093/nar/gkz333
Yarza P, Yilmaz P, Pruesse E, Glöckner FO, Ludwig W, Schleifer KH, Whitman WB, Euzéby J, Amann R, Rosselló-Móra R (2014) Uniting the classification of cultured and uncultured bacteria and archaea using 16S rRNA gene sequences. Nat Rev Microbiol 12:635–645. https://doi.org/10.1038/nrmicro3330
Yoon SH, Ha SM, Kwon S, Lim J, Kim Y, Seo H, Chun J (2017) Introducing EzBioCloud: a taxonomically united database of 16S rRNA gene sequences and whole-genome assemblies. Int J Syst Evol Microbiol 67:1613–1617. https://doi.org/10.1099/ijsem.0.001755
Acknowledgements
We are grateful to the graduate student (Yu-Jie Xin, Master’s degree) for her primary contribution to this study.
Funding
This work was financially supported by the National Natural Science Foundation of China (No. 32070003) and the National Science and Technology Fundamental Resources Investigation Program of China (No. 2021FY100900).
Author information
Authors and Affiliations
Contributions
YH, XM, ST, X-XL, MC performed the experiments and YH drafted the manuscript. JH and H-LC designed and supervised the study. JH and H-LC revised the manuscript. All authors of this article approved the submitted version.
Corresponding author
Ethics declarations
Conflict of interest
No competing interest exists.
Ethical approval
The article does not contain any study related to human participants or animals.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Hu, Y., Ma, X., Tan, S. et al. Genome-based classification of genera Halosegnis and Salella, and description of four novel halophilic archaea isolated from a tidal flat. Antonie van Leeuwenhoek 117, 51 (2024). https://doi.org/10.1007/s10482-024-01952-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s10482-024-01952-2