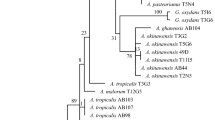
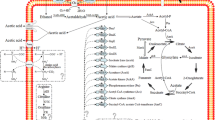
Abstract
Acetobacter senegalensis belongs to the group of acetic acid bacteria (AAB) that present potential biotechnological applications, for production of D-gluconate, cellulose and acetic acid. AAB can overcome heat and acid stresses by using strategies involving the overexpression of heat-shock proteins and enzymes from the complex pyrroquinoline-ADH, besides alcohol dehydrogenases (ADH). Nonetheless, the isolation of A. senegalensis and other AAB from food may be challenging due to presence of viable but non-culturable (VBNC) cells and due to uncertainties about nutritional requirements. To contribute for a better understanding of the ecology of AAB, this paper reports on the pangenome analysis of five strains of A. senegalensis recently isolated from a Brazilian spontaneous cocoa fermentation. The results showed biosynthetic clusters exclusively found in some cocoa-related AAB, such as those related to terpene pathways, which are important for flavour development. Genes related to oxidative stress were conserved in all the genomes, with multiple clusters. Moreover, there were genes coding for ADH and putative ABC transporters distributed in core, shell and cloud genomes, while chaperonin-encoding genes were present only in the core and soft-core genomes. Regarding quorum sensing, a response regulator gene was in the shell genome, and the gene encoding for acyl-homoserine lactone efflux protein was in the soft-core genome. There were quorum quenching-related genes, mainly encoding for lactonases, but also for acylases. Moreover, A. senegalensis did not have determinants of virulence or antibiotic resistance, which are good traits for strains intended to be applied in food fermentation.





Similar content being viewed by others
References
Akiko OK, Wang Y, Sachiko K et al (2002) Cloning and characterization of groESL operon in Acetobacter aceti. J Biosci Bioeng 94:140–147. https://doi.org/10.1016/S1389-1723(02)80134-7
Alcock BP, Raphenya AR, Lau TTY et al (2020) CARD 2020: antibiotic resistome surveillance with the comprehensive antibiotic resistance database. Nucleic Acids Res 48:D517–D525. https://doi.org/10.1093/nar/gkz935
Almeida OGG, De Martinis EC (2021) Metagenome-assembled genomes contributes to unravel the microbiome of cocoa fermentation. Appl Environ Microbiol. https://doi.org/10.1128/aem.00584-21
Almeida OGG, Pinto UM, Matos CB et al (2020) Does Quorum Sensing play a role in microbial shifts along spontaneous fermentation of cocoa beans? An in silico perspective. Food Res Int 131:109034. https://doi.org/10.1016/j.foodres.2020.109034
Almeida OGG, Vitulo N, De Martinis ECP, Felis GE (2021) Pangenome analyses of LuxS-coding genes and enzymatic repertoires in cocoa-related lactic acid bacteria. Genomics 113:1659–1670. https://doi.org/10.1016/j.ygeno.2021.04.010
Antipov D, Hartwick N, Shen M et al (2016) PlasmidSPAdes: assembling plasmids from whole genome sequencing data. Bioinformatics 32:3380–3387. https://doi.org/10.1093/bioinformatics/btw493
Aswini K, Gopal NO, Uthandi S (2020) Optimized culture conditions for bacterial cellulose production by Acetobacter senegalensis MA1. BMC Biotechnol 20:1–16. https://doi.org/10.1186/s12896-020-00639-6
Baker GC, Smith JJ, Cowan DA (2003) Review and re-analysis of domain-specific 16S primers. J Microbiol Methods 55:541–555. https://doi.org/10.1016/j.mimet.2003.08.009
Blin K, Shaw S, Kloosterman AM et al (2021) antiSMASH 6.0: improving cluster detection and comparison capabilities. Nucleic Acids Res 49:29–35. https://doi.org/10.1093/nar/gkab335
Brettin T, Davis JJ, Disz T et al (2015) RASTtk: a modular and extensible implementation of the RAST algorithm for building custom annotation pipelines and annotating batches of genomes. Sci Rep. https://doi.org/10.1038/srep08365
Bushnell B (2014) BBTools software package. https://jgi.doe.gov/data-and-tools/bbtools/
Carvalho CC, Phan NN, Chen Y, Reilly PJ (2015) Carbohydrate-binding module tribes. Biopolymers 103:203–214. https://doi.org/10.1002/bip.22584
Chakraborty S, Rani A, Dhillon A, Goyal A (2016) Polysaccharide Lyases. Elsevier, Amsterdam
Chen L, Zheng D, Liu B et al (2016) VFDB 2016: hierarchical and refined dataset for big data analysis—10 years on. Nucleic Acids Res 44:D694–D697. https://doi.org/10.1093/nar/gkv1239
Contreras-Moreira B, Vinuesa P (2013) GET_HOMOLOGUES, a versatile software package for scalable and robust microbial pangenome analysis. Appl Environ Microbiol 79:7696–7701. https://doi.org/10.1128/AEM.02411-13
De Roos J, De Vuyst L (2018) Acetic acid bacteria in fermented foods and beverages. CurrOpinBiotechnol 49:115–119. https://doi.org/10.1016/j.copbio.2017.08.007
De Roos J, Verce M, Weckx S, De Vuyst L (2020) Temporal Shotgun Metagenomics Revealed the Potential Metabolic Capabilities of Specific Microorganisms During Lambic Beer Production. Front Microbiol. https://doi.org/10.3389/fmicb.2020.01692
Doster E, Lakin SM, Dean CJ et al (2020) MEGARes 2.0: a database for classification of antimicrobial drug, biocide and metal resistance determinants in metagenomic sequence data. Nucleic Acids Res 48:D561–D569. https://doi.org/10.1093/nar/gkz1010
Esquivel-Elizondo S, Ilhan ZE, Garcia-Peña EI, Krajmalnik-Brown R (2017) Insights into Butyrate Production in a Controlled Fermentation System via Gene Predictions. mSystems. https://doi.org/10.1128/msystems.00051-17
Fang G, Rocha EPC, Danchin A (2008) Persistence drives gene clustering in bacterial genomes. BMC Genomics 9:4. https://doi.org/10.1186/1471-2164-9-4
Gao L, Wu X, Xia X, Jin Z (2021) Fine-tuning ethanol oxidation pathway enzymes and cofactor PQQ coordinates the conflict between fitness and acetic acid production by Acetobacter pasteurianus. MicrobBiotechnol 14:643–655. https://doi.org/10.1111/1751-7915.13703
Gautreau G, Bazin A, Gachet M et al (2020) PPanGGOLiN: depicting microbial diversity via a partitioned pangenome graph. PLoS Comput Biol 16:1–27. https://doi.org/10.1371/journal.pcbi.1007732
Gomes RJ, Borges MF, Rosa MF et al (2018) Acetic acid bacteria in the food industry: systematics, characteristics and applications. Food Technol Biotechnol 56:139–151. https://doi.org/10.17113/ftb.56.02.18.5593
Grandclément C, Tannières M, Moréra S et al (2015) Quorum quenching: role in nature and applied developments. FEMS Microbiol Rev 40:86–116. https://doi.org/10.1093/femsre/fuv038
Gupta SK, Padmanabhan BR, Diene SM et al (2014) ARG-annot, a new bioinformatic tool to discover antibiotic resistance genes in bacterial genomes. Antimicrob Agents Chemother 58:212–220. https://doi.org/10.1128/AAC.01310-13
Gurevich A, Saveliev V, Vyahhi N, Tesler G (2013) QUAST: quality assessment tool for genome assemblies. Bioinformatics 29:1072–1075. https://doi.org/10.1093/bioinformatics/btt086
Hammami R, Zouhir A, Ben Hamida J, Fliss I (2007) BACTIBASE: a new web-accessible database for bacteriocin characterization. BMC Microbiol 7:6–11. https://doi.org/10.1186/1471-2180-7-89
Huang L, Zhang H, Wu P et al (2018) DbCAN-seq: a database of carbohydrate-active enzyme (CAZyme) sequence and annotation. Nucleic Acids Res 46:D516–D521. https://doi.org/10.1093/nar/gkx894
Iida A, Ohnishi Y, Horinouchi S (2008) Control of acetic acid fermentation by quorum sensing via N-acylhomoserine lactones in Gluconacetobacter intermedius. J Bacteriol 190:2546–2555. https://doi.org/10.1128/JB.01698-07
Iida A, Ohnishi Y, Horinouchi S (2009) Identification and characterization of target genes of the GinI/GinR quorum-sensing system in Gluconacetobacter intermedius. Microbiology 155:3021–3032. https://doi.org/10.1099/mic.0.028613-0
Ishida T, Sugano Y, Shoda M (2002) Novel glycosyltransferase genes involved in the acetan biosynthesis of Acetobacter xylinum. Biochem Biophys Res Commun 295:230–235. https://doi.org/10.1016/S0006-291X(02)00663-0
Ishikawa M, Okamoto-Kainuma A, Jochi T et al (2010) Cloning and characterization of grpE in Acetobacter pasteurianus NBRC 3283. J Biosci Bioeng 109:25–31. https://doi.org/10.1016/j.jbiosc.2009.07.008
Jakob F, Quintero Y, Musacchio A et al (2019) Acetic acid bacteria encode two levan sucrase types of different ecological relationship. Environ Microbiol 21:4151–4165. https://doi.org/10.1111/1462-2920.14768
Koonin EV (2005) Orthologs, paralogs, and evolutionary genomics. Annu Rev Genet 39:309–338. https://doi.org/10.1146/annurev.genet.39.073003.114725
Koul S, Kalia VC (2017) Multiplicity of quorum quenching enzymes: a potential mechanism to limit quorum sensing bacterial population. Indian J Microbiol 57:100–108. https://doi.org/10.1007/s12088-016-0633-1
La China S, Zanichelli G, De Vero L, Gullo M (2018) Oxidative fermentations and exopolysaccharides production by acetic acid bacteria: a mini review. Biotechnol Lett 40:1289–1302. https://doi.org/10.1007/s10529-018-2591-7
Letunic I, Bork P (2019) Interactive Tree of Life (iTOL) v4: recent updates and new developments. Nucleic Acids Res 47:256–259. https://doi.org/10.1093/nar/gkz239
Levasseur A, Drula E, Lombard V et al (2013) Expansion of the enzymatic repertoire of the CAZy database to integrate auxiliary redox enzymes. Biotechnol Biofuels 6:1–14. https://doi.org/10.1186/1754-6834-6-41
Liu LP, Huang LH, Ding XT et al (2019) Identification of quorum-sensing molecules of N-acyl-homoserine lactone in Gluconacetobacter strains by liquid chromatography-tandem mass spectrometry. Molecules 24:2694. https://doi.org/10.3390/molecules24152694
Lynch KM, Zannini E, Wilkinson S et al (2019) Physiology of acetic acid bacteria and their role in vinegar and fermented beverages. Compr Rev Food Sci Food Saf 18:587–625. https://doi.org/10.1111/1541-4337.12440
Martínez-Núñez MA, López VEL (2016) Nonribosomal peptides synthetases and their applications in industry. Sustain Chem Process 4:1–8. https://doi.org/10.1186/s40508-016-0057-6
Matsutani M, Hirakawa H, Nishikura M et al (2011) Increased number of Arginine-based salt bridges contributes to the thermotolerance of thermotolerant acetic acid bacteria, Acetobacter tropicalis SKU1100. Biochem Biophys Res Commun 409:120–124. https://doi.org/10.1016/j.bbrc.2011.04.126
Nakamura AM, Nascimento AS, Polikarpov I (2017) Structural diversity of carbohydrate esterases. Biotechnol Res Innov 1:35–51. https://doi.org/10.1016/j.biori.2017.02.001
Nakano S, Fukaya M (2008) Analysis of proteins responsive to acetic acid in Acetobacter: Molecular mechanisms conferring acetic acid resistance in acetic acid bacteria. Int J Food Microbiol 125:54–59. https://doi.org/10.1016/j.ijfoodmicro.2007.05.015
Naseeb S, Ames RM, Delneri D, Lovell SC (2017) Rapid functional and evolutionary changes follow gene duplication in yeast. Proc R Soc B Biol Sci. https://doi.org/10.1098/rspb.2017.1393
Nayfach S, Shi ZJ, Seshadri R et al (2019) New insights from uncultivated genomes of the global human gut microbiome. Nature 568:505–510. https://doi.org/10.1038/s41586-019-1058-x
Ndoye B, Cleenwerck I, Engelbeen K et al (2007) Acetobacter senegalensis sp. nov., a thermotolerant acetic acid bacterium isolated in Senegal (sub-Saharan Africa) from mango fruit (Mangifera indica L.). Int J Syst Evol Microbiol 57:1576–1581. https://doi.org/10.1099/ijs.0.64678-0
Ng WL, Bassler BL (2009) Bacterial quorum-sensing network architectures. Annu Rev Genet 43:197–222. https://doi.org/10.1146/annurev-genet-102108-134304
Pal C, Bengtsson-Palme J, Rensing C et al (2014) BacMet: Antibacterial biocide and metal resistance genes database. Nucleic Acids Res 42:737–743. https://doi.org/10.1093/nar/gkt1252
Pelicaen R, Gonze D, Teusink B et al (2019) Genome-scale metabolic reconstruction of Acetobacter pasteurianus 386B, a candidate functional starter culture for cocoa bean fermentation. Front Microbiol. https://doi.org/10.3389/fmicb.2019.02801
Pritchard L, Glover RH, Humphris S et al (2016) Genomics and taxonomy in diagnostics for food security: soft-rotting enterobacterial plant pathogens. Anal Methods 8:12–24. https://doi.org/10.1039/c5ay02550h
Prjibelski A, Antipov D, Meleshko D et al (2020) Using SP Ades de novo assembler. Curr Protoc Bioinforma 70:1–29. https://doi.org/10.1002/cpbi.102
Qiu X, Zhang Y, Hong H (2021) Classification of acetic acid bacteria and their acid resistant mechanism. AMB Express 11:29. https://doi.org/10.1186/s13568-021-01189-6
Ruiz-Perez CA, Conrad RE, Konstantinidis KT (2021) MicrobeAnnotator: a user-friendly, comprehensive functional annotation pipeline for microbial genomes. BMC Bioinform 22:1–16. https://doi.org/10.1186/s12859-020-03940-5
Seemann T (2014) Prokka: rapid prokaryotic genome annotation. Bioinformatics 30:2068–2069. https://doi.org/10.1093/bioinformatics/btu153
Seemann T Abricate. https://github.com/tseemann/abricate
Shafiei R, Delvigne F, Babanezhad M, Thonart P (2013) Evaluation of viability and growth of Acetobacter senegalensis under different stress conditions. Int J Food Microbiol 163:204–213. https://doi.org/10.1016/j.ijfoodmicro.2013.03.011
Shafiei R, Zarmehrkhorshid R, Mounir M et al (2017) Influence of carbon sources on the viability and resuscitation of Acetobacter senegalensis during high-temperature gluconic acid fermentation. Bioprocess Biosyst Eng 40:769–780. https://doi.org/10.1007/s00449-017-1742-x
Shafiei R, Leprince P, Sombolestani AS et al (2019) Effect of sequential acclimation to various carbon sources on the proteome of Acetobacter senegalensis LMG 23690T and its tolerance to downstream process stresses. Front Microbiol 10:1–14. https://doi.org/10.3389/fmicb.2019.00608
Siedenburg G, Jendrossek D (2011) Squalene-hopene cyclases. Appl Environ Microbiol 77:3905–3915. https://doi.org/10.1128/AEM.00300-11
Simão FA, Waterhouse RM, Ioannidis P et al (2015) BUSCO: Assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 31:3210–3212. https://doi.org/10.1093/bioinformatics/btv351
Son H-J, Heo M-S, Kim Y-G, Lee S-J (2001) Optimization of fermentation conditions for the production of bacterial cellulose by a newly isolated Acetobacter sp.A9 in shaking cultures. Biotechnol Appl Biochem 33:1. https://doi.org/10.1042/ba20000065
Subramoni S, Venturi V (2009) LuxR-family “solos”: Bachelor sensors/regulators of signalling molecules. Microbiology 155:1377–1385. https://doi.org/10.1099/mic.0.026849-0
Tran TN, Phong HX, ThaoAnh BT et al (2018) High-temperature production of acerola vinegar using thermotolerant Acetobacter senegalensis A28. Vietnam J Sci Technol Eng 60:13–18. https://doi.org/10.31276/vjste.60(3).13
Trček J, Dogsa I, Accetto T, Stopar D (2021) Acetan and acetan-like polysaccharides: genetics, biosynthesis, structure, and viscoelasticity. Polymers (basel) 13:1–16. https://doi.org/10.3390/polym13050815
Valera MJ, Mas A, Streit WR, Mateo E (2016) GqqA, a novel protein in Komagataeibacter europaeus involved in bacterial quorum quenching and cellulose formation. Microb Cell Fact 15:1–15. https://doi.org/10.1186/s12934-016-0482-y
Versalovic J, Schneider M, De Bruijn FJ, Lupski J (1994) Genomic fingerprinting of bacteria using repetitive sequence-based polymerase chain reaction. Methods Mol Cell Biol 5:25–40
Vinuesa P, Ochoa-Sánchez LE, Contreras-Moreira B (2018) GET_PHYLOMARKERS, a software package to select optimal orthologous clusters for phylogenomics and inferring pan-genome phylogenies, used for a critical geno-taxonomic revision of the genus Stenotrophomonas. Front Microbiol 9:1–22. https://doi.org/10.3389/fmicb.2018.00771
Wang B, Shao Y, Chen T et al (2015a) Global insights into acetic acid resistance mechanisms and genetic stability of Acetobacter pasteurianus strains by comparative genomics. Sci Rep 5:1–14. https://doi.org/10.1038/srep18330
Wang Z, Zang N, Shi J et al (2015b) Comparative proteome of Acetobacter pasteurianus Ab3 during the high acidity rice vinegar fermentation. Appl Biochem Biotechnol 177:1573–1588. https://doi.org/10.1007/s12010-015-1838-1
Wu JJ, Ma YK, Zhang FF, Chen FS (2012) Biodiversity of yeasts, lactic acid bacteria and acetic acid bacteria in the fermentation of “Shanxi aged vinegar”, a traditional Chinese vinegar. Food Microbiol 30:289–297. https://doi.org/10.1016/j.fm.2011.08.010
Xia K, Zang N, Zhang J et al (2016) New insights into the mechanisms of acetic acid resistance in Acetobacter pasteurianus using iTRAQ-dependent quantitative proteomic analysis. Int J Food Microbiol 238:241–251. https://doi.org/10.1016/j.ijfoodmicro.2016.09.016
Xia K, Han C, Xu J, Liang X (2020) Transcriptome response of Acetobacter pasteurianus Ab3 to high acetic acid stress during vinegar production. Appl Microbiol Biotechnol 104:10585–10599. https://doi.org/10.1007/s00253-020-10995-0
Yang H, Yu Y, Fu C, Chen F (2019) Bacterial acid resistance toward organic weak acid revealed by RNA-seq transcriptomic analysis in Acetobacter pasteurianus. Front Microbiol 10:1–14. https://doi.org/10.3389/fmicb.2019.01616
Yin H, Zhang R, Xia M et al (2017) Effect of aspartic acid and glutamate on metabolism and acid stress resistance of Acetobacter pasteurianus. Microb Cell Fact 16:1–14. https://doi.org/10.1186/s12934-017-0717-6
Zankari E, Hasman H, Cosentino S et al (2012) Identification of acquired antimicrobial resistance genes. J Antimicrob Chemother 67:2640–2644. https://doi.org/10.1093/jac/dks261
Zhang Z, Ma H, Yang Y et al (2015) Protein profile of Acetobacter pasteurianus HSZ3-21. CurrMicrobiol 70:724–729. https://doi.org/10.1007/s00284-015-0777-y
Acknowledgements
The authors are thankful to the São Paulo Research Foundation (FAPESP)–Brazil for a research grant (process 2018/13564-3). FAPESP is especially acknowledged by O.G.G.A. for a Ph.D. fellowship (process 2017/137596). This study was financed in part by the Coordenação de Aperfeiçoamento de Pessoal de Nível Superior–Brazil (CAPES; finance code 001). E.C.P.D.M. is a research fellow of the National Council for Scientific and Technological Development (CNPq)–Brazil.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors have no competing interests to disclose.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Almeida, O.G.G., Gimenez, M.P. & De Martinis, E.C.P. Comparative pangenomic analyses and biotechnological potential of cocoa-related Acetobacter senegalensis strains. Antonie van Leeuwenhoek 115, 111–123 (2022). https://doi.org/10.1007/s10482-021-01684-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10482-021-01684-7