Abstract
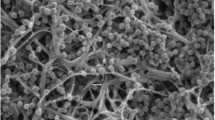
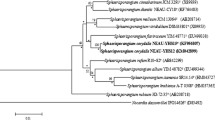
A Gram-stain positive actinomycete, strain AZ1-19T, isolated from roots of Azadirachta indica A. Juss. var. siamensis Valeton, collected from Chachoengsao province, Thailand, was characterised taxonomically by using a polyphasic approach. Strain AZ1-19T was found to have characteristics consistent with those of members of the genus Micromonospora. The cell wall peptidoglycan of the strain was found to contain meso-diaminopimelic acid. The predominant phospholipids were identified as diphosphatidylglycerol, phosphatidylethanolamine and phosphatidylinositol. The characteristic whole-cell sugars were identified as glucose, xylose, galactose and mannose. The major menaquinones were found to be MK-9(H6), MK-10(H6) and MK-10(H8), and the major cellular fatty acids were identified as iso-C16:0, iso-C15:0, anteiso-C15:0 and anteiso-C17:0. Comparative analysis of 16S rRNA gene sequences revealed that strain AZ1-19T is closely related to Micromonospora costi CS1-12T (98.75% similarity), Micromonospora avicenniae 268506T (98.75%), Micromonospora haikouensis 232617T (98.68%) and Micromonospora siamensis TT2-4T (98.61%), whilst the corresponding phylogenetic analysis based on partial gyrase subunit B (gyrB) gene sequences indicated that strain AZ1-19T forms a clade with M. avicenniae 268506T with a high bootstrap value. The DNA G + C content was determined to be 69.8 mol%. Moreover, a combination of DNA–DNA relatedness values and some phenotypic and chemotaxonomic properties indicated that the strain could be distinguished from closely related species. Therefore, it is considered that strain AZ1-19T represents a novel Micromonospora species for which the name Micromonospora azadirachtae sp. nov. is proposed. The type strain is AZ1-19T (= KCTC 39941T = NBRC 112784T = JCM 32148T = TISTR 2559T).



Similar content being viewed by others
References
Carro L, Pukall R, Spröer C, Kroppenstedt RM, Trujillo ME (2012) Micromonospora cremea sp. nov. and Micromonospora zamorensis sp. nov., isolated from the rhizosphere of Pisum sativum. Int J Syst Evol Microbiol 62:2971–2977
Collins MD, Pirouz T, Goodfellow M, Minnikin DE (1977) Distribution of menaquinones in actinomycetes and corynebacteria. J Gen Microbiol 100:221–230
Coombs JT, Franco CMM (2003) Visualization of an endophytic Streptomyces species in wheat seed. Appl Environ Microbiol 69:4260–4262
Ezaki T, Hashimoto Y, Yabuuchi E (1989) Fluorometric deoxyribonucleic acid-deoxyribonucleic acid hybridization in microdilution wells as an alternative to membrane filter hybridization in which radioisotopes are used to determine genetic relatedness among bacterial strains. Int J Syst Bacteriol 39:224–229
Felsenstein J (1981) Evolutionary trees from DNA sequences: a maximum likelihood approach. J Mol Evol 17:368–376
Felsenstein J (1985) Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783–791
Fitch WM (1971) Toward defining the course of evolution: minimum change for specific tree topology. Syst Zool 20:406–416
Garcia LC, Martínez-Molina E, Trujillo ME (2010) Micromonospora pisi sp. nov., isolated from root nodules of Pisum sativum. Int J Syst Evol Microbiol 60:331–337
Genilloud O (2012) Genus I Micromonospora Ørskov 1923 156AL. In: Goodfellow M, Kämpfer P, Busse HJ, Trujillo ME, Suzuki K, Ludwig W, Whitman WB (eds) Bergey’s manual of systematic bacteriology, vol 5. Springer, New York, pp 1039–1057
Hall TA (1999) BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98 NT. Nucleic Acids Symp Ser 41:95–98
Hasegawa S, Meguro A, Shimizu M, Nishimura T, Kunoh H (2006) Endophytic actinomycetes and their interactions with host plants. Actinomycetologica 20:72–81
Igarashi Y, Trujillo ME, Martínez-Molina E et al (2007) Antitumor anthraquinones from an endophytic actinomycete Micromonospora lupini sp. nov. Bioorg Med Chem Lett 17:3702–3705
Igarashi Y, Yanase S, Suigamoto K et al (2011a) Lupinacidin C, an inhibitor of tumour cell invasion from Micromonospora lupini. J Nat Prod 74:862–865
Igarashi Y, Ogura H, Furihata K et al (2011b) Maklamicin, an antibacterial polyketide from an endophytic Micromonospora sp. J Nat Prod 74:670–674
Jacobson E, Grauville WC, Fogs CE (1958) Colour harmony manual 4th. Container Corporation of America, Chicago
Kawamoto I (1989) Genus Micromonospora Ørskov 1923, 147AL. In: Williams ST, Sharpe ME, Holt JG (eds) Bergey’s Manual of Systematic Bacteriology, vol 4. Williams & Wilkins, Baltimore, pp 2442–2450
Kimura M (1980) A simple method for estimating evolutionary rate of base substitutions through comparative studies of nucleotide sequences. J Mol Evol 16:111–120
Kirby BM, Meyers PR (2010) Micromonospora tulbaghiae sp. nov., isolated from the leaves of wild garlic, Tulbaghia violacea. Int J Syst Evol Microbiol 60:1328–1333
Kittiwongwattana C, Thanaboripat D, Laosinwattana C, Koohakan P, Parinthawong N, Thawai C (2015) Micromonospora oryzae sp. nov., isolated from roots of upland rice. Int J Syst Evol Microbiol 65:3818–3823
Kroppenstedt RM (1985) Fatty acid and menaquinone analysis of actinomycetes and related organisms. In: Goodfellow M, Minnikin DE (eds) Chemical methods in bacterial systematics, vol 20. Academic Press, New York, pp 173–199
Kudo T, Matsushima K, Itoh T, Sasaki J, Suzuki K (1998) Description of four new species of the genus Kineosporia: Kineosporia succinea sp. nov., Kineosporia rhizophila sp. nov., Kineosporia mikuniensis sp. nov. and Kineosporia rhamnosa sp. nov., isolated from plant samples, and amended description of the genus Kineosporia. Int J Syst Bacteriol 48:1245–1255
Kumar S, Stecher G, Tamura K (2016) MEGA 7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 33(7):1870–1874
Kuncharoen N, Pittayakhajonwut P, Tanasupawat S (2018) Micromonospora globbae sp. nov., an endophytic actinomycete isolated from roots of Globba winitii C. H. Wright. Int J Syst Evol Microbiol 68:1073–1077
Lane DJ (1991) 16S/23S rRNA sequencing. In: Stackebrandt E, Goodfellow M (eds) Nucleic acid techniques in bacterial systematics. Wiley, Chichester, pp 115–148
Lechevalier MP, De Bièvre C, Lechevalier HA (1977) Chemotaxonomy of aerobic actinomycetes: phospholipid composition. Biochem Syst Ecol 5:249–260
Li L, Mao YJ, Xie QY, Deng Z, Hong K (2013) Micromonospora avicenniae sp. nov., isolated from a root of Avicennia marina. Antonie Van Leeuwenhoek 103:1089–1096
Mikami H, Ishida Y (1983) Post-column fluorometric detection of reducing sugar in high-performance liquid chromatography using arginine. Bunseki Kagaku 32:E207–E210
Minnikin DE, O’Donnell AG, Goodfellow M, Alderson G, Athalye M, Schaal A, Parlett JH (1984) An integrated procedure for the extraction of bacterial isoprenoid quinones and polar lipids. J Microbiol Methods 2:233–241
Okazaki T (2003) Studies on actinomycetes isolated from plant leaves. In: Kurtboke I, Hayakawa M, Terekhova L, Okazaki T (eds) Selective isolation of actinomycetes. National Library of Australia, Queensland, pp 102–122
Ørskov J (1923) Investigations into the morphology of the ray fungi. Levin and Munksgaard, Copenhagen
Phongsopitanun P, Kudo T, Mori M, Shiomi K, Pittayakhajonwut P, Suwanborirux K, Tanasupawat S (2015) Micromonospora fluostatini sp. nov., isolated from marine sediment. Int J Syst Evol Microbiol 65:4417–4423
Qin S, Li J, Chen H-H, Zhao G-Z, Zhu W-Y, Jiang C-L, Xu L-H, Li W-J (2009) Isolation, diversity, and antimicrobial activity of rare actinobacteria from medicinal plants of tropical rain forests in Xinshuangbanna, China. Appl Environ Microbiol 75:6176–6186
Rosselló-Móra R, Trujillo ME, Sutcliffe IC (2017) Introducing a digital protologue: a timely move towards a database-driven systematics of archaea and bacteria. Antonie Van Leeuwenhoek 110:455–456
Saitou N, Miura K (1963) Preparation of transforming deoxyribonucleic acid by phenol treatment. Biochim Biophys Acta 72:619–629
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425
Sasser M (1990) Identification of bacteria by gas chromatography of cellular fatty acids. Technical Note 101. MIDI, Newark
Shirling EB, Gottlieb D (1966) Methods for characterization of Streptomyces species. Int J Syst Bacteriol 16:313–340
Staneck JL, Robert GD (1974) Simplified approach to identification of aerobic actinomycetes by thin-layer chromatography. Appl Microbiol 28:226–231
Supong K, Suriyachadkun C, Pittayakhajonwut P, Suwanborirux K, Thawai C (2013) Micromonospora spongicola sp. nov., an actinomycete isolated from a marine sponge in the gulf of Thailand. J Antibiot 66:505–509
Suriyachadkun C, Chunhametha S, Thawai C, Tamura T, Potacharoen W, Kirtikara K, Sanglier J (2009) Planotetraspora thailandica sp. nov., isolated from soil in Thailand. Int J Syst Evol Microbiol 59:992–997
Tamaoka J (1994) Determination of DNA base composition. In: Goodfellow M, O’Donnell AG (eds) Chemical methods in prokaryotic systematics. Wiley, Chichester, pp 463–470
Tamaoka J, Komagata K (1984) Determination of DNA base composition by reversed-phase high-performance liquid chromatography. FEMS Microbiol Lett 25:125–128
Tamaoka J, Katayama-Fujimura Y, Kuraishi H (1983) Analysis of bacterial menaquinone mixtures by high performance liquid chromatography. J Appl Bacteriol 54:31–36
Tanasupawat S, Jongrungruangchok S, Kudo T (2010) Micromonospora marina sp. nov., isolated from sea sand. Int J Syst Evol Microbiol 60:648–652
Thawai C (2015) Micromonospora costi sp. nov., isolated from a leaf of Costus speciosus. Int J Syst Evol Microbiol 65:1456–1461
Thawai C, Tanasupawat S, Itoh T, Suwanborirux K, Kudo T (2005) Micromonospora siamensis sp. nov., isolated from Thai peat swamp forest. J Gen Appl Microbiol 51:229–234
Tomiyasu I (1982) Mycolic acid composition and thermally adaptative changes in Nocardia asteroids. J Bacteriol 151:828–837
Trujillo ME, Kroppenstedt RM, Schumann P, Carro L, Martínez-Molina E (2006) Micromonospora coriariae sp. nov., isolated from root nodules of Coriaria myrtifolia. Int J Syst Evol Microbiol 56:2381–2385
Uchida K, Aida K (1984) An improved method for the glycolate test for simple identification of the acyl type of bacterial cell walls. J Gen Appl Microbiol 30:131–134
Wang C, Xu XX, Qu Z, Wang HL, Lin HP, Xie QY, Ruan JS, Hong K (2011) Micromonospora rhizosphaerae sp. nov., isolated from mangrove rhizosphere soil. Int J Syst Evol Microbiol 61:320–324
Wayne LG, Brenner DJ, Colwell RR, Grimont PAD, Kandler O, Krichevsky MI, Moore LH, Moore WEC, Murray RGE et al (1987) International committee on systematic bacteriology. Report of the ad hoc committee on the reconciliation of approaches to bacterial systematic. Int J Syst Bacteriol 37:463–464
Xie QY, Qu Z, Lin HP, Li L, Hong K (2012) Micromonospora haikouensis sp. nov., isolated from mangrove soil. Antonie Van Leeuwenhoek 101:649–655
Yoon SH, Ha SM, Kwon S, Lim J, Kim Y, Seo H, Chun J (2017) Introducing EzBiocloud: a taxonomically united database of 16S rRNA and whole genome assemblies. Int J Syst Evol Microbiol 67:1613–1617
Acknowledgements
We would like to thank Dr A. Matsumoto and Prof Dr Y. Takahashi, Department of Drug Discovery, Kitasato University for the isoprenoid quinone analysis by LC-MS, and appreciate Assoc Prof Dr C. Thawai, Department of Biology, Faculty of Science, King’s Mongkut Institute of Technology Ladkrabang, for providing M. costi CS1-12T. We also thank the 2015 Royal Golden Jubilee Ph. D. Programme scholarship to N. K. for financial support under the Thailand Research Fund.
Author information
Authors and Affiliations
Contributions
We declare that the present study was performed by the authors named in this article. N. Kuncharoen, S. Tanasupawat designed the study; N. Kuncharoen, S. Tanasupawat, and T. Kudo performed experiments on isolation, identification, analysis, and interpretation; M. Ohkuma gave technical support and conceptual advice. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflicts of interest
The authors declare that there are no conflicts of interest.
Ethical approval
This article does not contain any studies with human participants and/or animals performed by any of the authors. The formal consent is not required in this study.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Kuncharoen, N., Kudo, T., Ohkuma, M. et al. Micromonospora azadirachtae sp. nov., isolated from roots of Azadirachta indica A. Juss. var. siamensis Valeton. Antonie van Leeuwenhoek 112, 253–262 (2019). https://doi.org/10.1007/s10482-018-1152-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10482-018-1152-3