Abstract
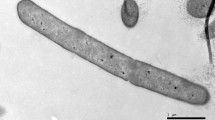
A Gram-negative strain, designated SA1T, was isolated from a sand specimen from Yangyang Beach, South Korea. The cells of SA1T were observed to be red pigmented, strictly aerobic, non-motile rods. Strain SA1T was found to grow optimally at 30 °C and pH 7 (pH growth range, pH 7–9). Phylogenetic analyses of the 16S rRNA gene sequence of SA1T showed that the organism belongs to the phylum Deinococcus-Thermus and forms a branch within the genus Deinococcus, with Deinococcus actinosclerus BM2T as a close relative (99.8 % sequence similarity). The strain was found to be catalase positive and oxidase negative and to metabolise 2-ketogluconate, acetate, d,l-3-hydroxybutyrate, d-glucose, d-maltose, d-mannitol, d-mannose, d-melibiose, d-sorbitol, d-sucrose, glycogen, l-alanine, l-histidine, l-malate, l-proline and n-valerate. The guanine plus cytosine (G + C) base composition of the DNA was 69.5 mol %, as determined by the thermal denaturation method. The predominant respiratory quinone was identified as menaquinone with eight isoprene units (MK-8). The polar lipid profile of strain SA1T showed the presence of several unidentified glycolipids, phosphoglycolipids, phospholipids, aminophospholipids and unidentified lipids. In addition, the most abundant fatty acids of strain SA1T were identified as C15:1 ω6c, C16:1 ω7c and C17:0. On the basis of phylogenetic analyses, DNA–DNA hybridisation and biochemical characteristics, strain SA1T (=KCTC 33741T = JCM 31047T) is concluded to represent a novel species of the genus Deinococcus, for which the name Deinococcus arenae sp. nov. is proposed.

Similar content being viewed by others
References
Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25:3389–3402
Brooks BW, Murray RGE (1981) Nomenclature for “Micrococcus radiodurans” and other radiation-resistant cocci: Deinococcaceae fam. nov. and Deinococcus gen. nov., including five species. Int J Syst Evol Microbiol 31:353–360
Buck JD (1982) Nonstaining (KOH) method for determination of Gram reactions of marine bacteria. Appl Environ Microbiol 44:992–993
Callegan RP, Nobre MF, McTernan PM, Battista JR, Navarro-González R, McKay CP, da Costa MS, Rainey FA (2008) Description of four novel psychrophilic, ionizing radiation-sensitive Deinococcus species from alpine environments. Int J Syst Evol Microbiol 58:1252–1258
Cha S, Srinivasan S, Seo T, Kim MK (2014a) Deinococcus radiotolerans sp. nov., a gamma-radiationresistant bacterium isolated from gamma ray-irradiated soil. Antonie Van Leeuwenhoek 105:229–235
Cha S, Srinivasan S, Seo T, Kim MK (2014b) Deinococcus soli sp. nov., a Gamma-Radiation-Resistant Bacterium Isolated from Rice Field Soil. Curr Microbiol 68:777–783
Collins MD, Jones D (1981) Distribution of isoprenoid quinone structural types in bacteria and their taxonomic implication. Microbiol Rev 45:316–354
De Ley J, Cattoir JH, Reynaerts A (1970) The quantitative measurement of DNA hybridization from renaturation rates. Eur J Biochem 12:133–142
Felsenstein J (1985) Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783–791
Fitch WM (1971) Toward defining the course of evolution: minimum change for a specified tree topology. Syst Zool 20:406–416
Gillis M, De Ley J, De Cleene M (1970) The determination of molecular weight of bacterial genome DNA from renaturation rates. Eur J Biochm 12:143–153
Gonzalez JM, Saiz-Jimenez C (2002) A fluorimetric method for the estimation of G+C mol % content in microorganisms by thermal denaturation temperature. Environ Microbiol 4:770–773
Hiraishi A, Ueda Y, Ishihara J, Mori T (1996) Comparative lipoquinone analysis of influent sewage and activated sludge by highperformance liquid chromatography and photodiode array detection. J Gen Appl Microbiol 42:457–469
Hirsch P, Gallikowski CA, Siebert J, Peissl K, Kroppenstedt R, Schumann P, Stackebrandt E, Anderson R (2004) Deinococcus frigens sp. nov., Deinococcus saxicola sp. nov., and Deinococcus marmoris sp. nov., low temperature and draught-tolerating, UV-resistant bacteria from continental Antarctica. Syst Appl Microbiol 27:636–645
Im WT, Jung HM, Ten LN, Kim MK, Bora N, Goodfellow M, Lim S, Jung J, Lee ST (2008) Deinococcus aquaticus sp. nov., isolated from fresh water, and Deinococcus caeni sp. nov., isolated from activated sludge. Int J Syst Evol Microbiol 58:2348–2353
Jit S, Dhadwal M, Prakash O, Lal R (2008) Flavobacterium lindanitolerans sp. nov., isolated from hexachlorocyclohexane-contaminated soil. Int J Syst Evol Microbiol 58:1665–1669
Joo ES, Lee JJ, Kang MS, Lim S, Jeong SW, Kim EB, Jeon SH, Srinivasan S, Kim MK (2015) Deinococcus actinosclerus sp. nov., a novel bacterium isolated from soil, South Korea. Int J Syst Evol Microbiol. doi:10.1099/ijsem.0.000825
Kim OS, Cho YJ, Lee K, Yoon SH, Kim M, Na H, Park SC, Jeon YS, Lee JH, Yi H, Won S, Chun J (2012) Introducing EzTaxon-e: a prokaryotic 16S rRNA gene sequence database with phylotypes that represent uncultured species. Int J Syst Evol Microbiol 62:716–721
Kimura M (1980) A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol 16:111–120
Komagata K, Suzuki KI (1987) Lipid and cell-wall analysis in bacterial systematics. Methods Microbiol 19:161–205
Kuykendall LD, Roy MA, O’Neill JJ, Devine TE (1988) Fatty acids, antibiotic resistance and deoxyribonucleic acid homology groups of Bradyrhizobium japonicum. Int J Syst Evol Microbiol 38:358–361
Loveland-Curtze J, Vanya IM, Jean EB (2011) Evaluation of a new fluorimetric DNA–DNA hybridzation method. Can J Microbiol 57:250–255
Minnikin DE, O’Donnell AG, Goodfellow M, Alderson G, Athalye M, Schaal A, Parlett JH (1984) An integrated procedure for the extraction of bacterial isoprenoid quinones and polar lipids. J Microbiol Methods 2:233–241
Peng F, Zhang L, Luo X, Dai J, An H, Tang Y, Fang C (2009) Deinococcus xinjiangensis sp. nov., isolated from desert soil. Int J Syst Evol Microbiol 59:709–713
Rainey FA, Ray K, Ferreira M, Gatz BZ, Nobre MF, Bagaley D, Rash BA, Park MJ, Earl AM, Shank NC, Small AM, Henk MC, Battista JR, Kämpfer P, da Costa MS (2005) Extensive diversity of ionizing-radiation-resistant bacteria recovered from Sonoran Desert soil and description of nine new species of the genus Deinococcus obtained from a single soil sample. Appl Environ Microbiol 71:5225–5235
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425
Srinivasan S, Kim MK, Lim S, Joe M, Lee M (2012a) Deinococcus daejeonensis sp. nov., isolated from sludge in a sewage disposal plant. Int J Syst Evol Microbiol 62:1265–1270
Srinivasan S, Lee JJ, Lim S, Joe M, Kim MK (2012b) Deinococcus humi sp. nov., isolated from soil. Int J Syst Evol Microbiol 62:2844–2850
Stackebrandt E, Goebel BM (1994) Taxonomic note: a place for DNA–DNA reassociation and 16S rRNA sequence analysis in the present species definition in bacteriology. Int J Syst Evol Microbiol 44:846–849
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28:2731–2739
Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 25:4876–4882
Wayne LG, Brenner DJ, Colwell RR, Grimont PAD, Kandler O, Krichevsky MI, Moore LH, Moore WEC, Murray RGE, Stackebrandt E, Starr MP, Truper HG (1987) International Committee on Systematic Bacteriology. Report of the ad hoc committee on reconciliation of approaches to bacterial systematics. Int J Syst Bacteriol 37:463–464
Weisburg WG, Barns SM, Pelletier DA, Lane DJ (1991) 16S ribosomal DNA amplification for phylogenetic study. J Bacteriol 173:697–703
White O, Eisen JA, Heidelberg JF, Hickey EK, Peterson JD, Dodson RJ, Haft DH, Gwinn ML, Nelson WC, Richardson DL, Moffat KS, Qin H, Jiang L, Pamphile W, Crosby M, Shen M, Vamathevan JJ, Lam P, McDonald L, Utterback T, Zalewski C, Makarova KS, Aravind L, Daly MJ, Minton KW, Fleischmann RD, Ketchum KA, Nelson KE, Salzberg S, Smith HO, Venter JC, Fraser CM (1999) Genome sequence of the radioresistant bacterium Deinococcus radiodurans R1. Science 286:1571–1577
Yoo SH, Weon HY, Kim SJ, Kim YS, Kim BY, Kwon SW (2010) Deinococcus aerolatus sp. nov. and Deinococcus aerophilus sp. nov., isolated from air samples. Int J Syst Evol Microbiol 60:1191–1195
Zhang L, Qin BF, Wang Y, Fang CX (2011) Deinococcus soli sp. nov., a gamma- and uv-radiation-resistant bacterium from north-west China. Mikrobiologiia 80:818–825
Acknowledgments
We are grateful to Dr. Bernhard Schink for help with etymology, and to Dr. Myung Kyum Kim for providing D. actinosclerus BM2T. This research was supported by National Institute of Environmental Research, Republic of Korea.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
There are no conflicts of interest to declare.
Additional information
Dongwook Lee, Seho Cha contributed equally to this work.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Lee, D., Cha, S., Jang, J.H. et al. Deinococcus arenae sp. nov., a novel species isolated from sand in South Korea. Antonie van Leeuwenhoek 109, 1055–1062 (2016). https://doi.org/10.1007/s10482-016-0705-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10482-016-0705-6