Abstract
Sansevieria species are valued in Europe as potted houseplants because of their modest maintenance requirements and low susceptibility to diseases and pests. Water-soaked leaf spots that eventually coalesced into large, drying lesions were observed on Sansevieria trifasciata freshly imported from Costa Rica. A Colletotrichum was isolated from the fungal fruiting bodies that developed on these necroses. It was confidently determined to be C. sansevieriae based on the host plant and microbiological as well as molecular biology test results. This is the first detection of C. sansevieriae in Germany.
Similar content being viewed by others
Introduction
Species of Sansevieria occur naturally in Africa, the Arabian Peninsula (Yemen), India, Sri Lanka, Myanmar and the Comoros. They are xerophytic, perennial, herbaceous, caulescent or acaulescent plants, sometimes branching at the base and spreading by underground rhizomes or above-ground stolons. The leaves, flat or cylindrical, arranged in rosettes, are leathery to succulent (Newton 2020). Currently about 85 species are accepted by Newton (2020), whom we follow in this work in terms of retaining the genus name Sansevieria, although recent phylogenetic studies suggest that Sansevieria might represent a clade of herbaceous plants within the genus Dracaena only (Takawira-Nyenya et al. 2018; van Kleinwee et al. 2022).
Used as fiber crop (Brink and Achigan-Dako 2012; Fiscal and Dandan 2016; Adeniyi et al. 2020) or for ethno–botanical purposes (Mohana et al. 2008; Takawira-Nyenya and Stedje 2011; Takawira-Nyenya et al. 2014) Sansevieria is of regional economic significance. Moreover, especially varieties of S. trifasciata are worldwide cultivated as ornamentals. They are used outdoors in subtropical and tropical regions as ground cover or elements of landscaping, while in cooler regions indoors as potted houseplant and for decorative purposes (Henley 1982; Nakamura et al. 2006). Their advantages are modest maintenance, especially in water and nutrient supply, and low susceptibility to pests and diseases.
However, several fungal pathogens are recognized causing leaf spots on Sansevieria spp., such as Chaetomella sp. (Li et al. 2013), Stemphylium lycopersici (Kee et al. 2017a), S. vesicarium (Ahmadpour and Poursafar 2018), Neoscytalidium dimidiatum (Kee et al. 2017b; Monteles et al. 2020), Lasiodiplodia spp. (Kee et al. 2019), Curvularia spp. (Kee et al. 2020a), Fusarium spp. (Kee et al. 2020b) as well as Colletotrichum neosansevieriae and Stachybotrys sansevieriicola (Crous et al. 2015). However, based on the number of reports, another fungal leaf spot pathogen, appears to be of greater importance: After observations of leaf spot (anthracnose) symptoms on S. trifasciata ‘Laurentii’ occurring since 1996 in subtropical regions of Japan, a Colletotrichum species was found as causal agent and introduced as new species, C. sansevieriae (Nakamura et al. 2006). Subsequently, this pathogen was reported from other subtropical and tropical regions like Victoria/Australia, Florida/USA, Madhya Pradesh/India, Costa Rica, Korea, Iran, Thailand, and Malaysia (Aldaoud et al. 2011; Campoverde and Palmateer 2012; Gautam et al. 2012; Palmateer et al. 2012; Karimi et al. 2017; Kee et al. 2020c; Li et al. 2020; Park et al. 2013; Pérez-León et al. 2013). In all reported cases, the infected host species was S. trifasciata.
Phylogenetical studies revealed that C. sansevieriae belongs to the clade ‘Sansevieriae’ which comprises species that infect succulent plants originating from Africa (Nakamura et al. 2018).
In spring 2022, leaf spots were observed in several cultivars of Sansevieria trifasciata (‘Robusta’, ‘Zeylanica’, ‘Aubrytiana Sayuri’, ‘Futura’, ‘Laurentii’, and ‘Moonshine’) at an ornamental plant nursery in Lower Saxony, Germany. The plants were imported from Costa Rica in February 2022. In order to be able to take appropriate countermeasures, inhibit further spread and prevent repeated occurrence, investigations were conducted to determine the cause of this disease.
Material and Methods
Source of Samples
In April 2022, several symptomatic specimens of the cultivars ‘Laurentii’ and ‘Moonshine’ were brought from the above-mentioned nursery to the laboratory of the Plant Protection Office of the Chamber of Agriculture of Lower Saxony, Oldenburg.
Microbiological Identification
After visual inspection of the plants, detached symptomatic leaves were incubated in humid chambers on wet filter paper at room temperature.
Under axenic conditions, without surface disinfection, small sections of leave tissue taken from the border of leaf spots were laid out on half strength potato dextrose agar (PDA50%; 19.5 g l−1 potato extract glucose agar (Carl Roth, Karlsruhe, Germany), 7.5 g l−1 agar) and incubated at room temperature. Single spore isolates were produced on PDA50% by spreading a conidial suspension with a Drigalski spatula successively onto several plates, and finally separating solitary microcolonies.
Mycelia plugs (5 mm diameter) were transferred to PDA50% and the irregularly roundish grown colonies were measured along the axis of widest expansion after 14 days of growth at 25 °C in the dark (n = 10).
Conidia were harvested from mature fruiting bodies on diseased plant tissue, suspended in water and examined microscopically (Eclipse Ni‑U; Nikon Europe B.V., Amstelveen, NL). Appressoria were obtained using the slide culture method described by Smith and Black (1990), and the length was measured.
Microscopic images and measurements of characteristic structures such as conidia (n = 200) and appressoria (n = 50) were made using the digital camera DS-Fi3, equipped with the corresponding software NIS-Elements D 5.20.01 (Nikon Europe B.V., Amstelveen, NL). All measurements were made at 400 × magnification and are reported as (minimum–) mean ± standard deviation (–maximum) and as median.
Phylogenetic Identification
For DNA isolation freshly grown mycelium was scraped off in a 2 ml reaction tube and slightly homogenized with a pestle prior to further processing. DNA was extracted using the innuPREP Plant DNA Kit (Analytik Jena AG, Jena, Germany) following the manufacturer’s instructions given in protocol 1.
Internal transcribed spacer (ITS) gene regions were amplified by using primer pairs ITS1F/ITS4 (Gardes and Bruns 1993; White et al. 1990) were used. PCR was performed in a reaction volume of 50 µl containing final concentrations of 1 × ready-to-use MyTaq Plant-PCR Mix (Bioline Meridian Bioscience, London, UK), 5 µl genomic DNA (1:10 diluted) and 0.4 µM of each primer. Amplification conditions were 5 min at 95 °C, followed by 30 cycles at 94 °C for 30 s, 55 °C for 45 s, 72 °C for 90 s, followed by a final extension at 72 °C for 10 min.
PCR products were purified using innuPREP PCRpure Kit (Analytik Jena AG, Jena, Germany). For bidirectional Sanger sequencing the same primer pairs were used as well. A consensus sequences was prepared by assembling and analyzing electropherograms in DNA Sequence Assembler v5 (Heracle BioSoft S.R.L, Arges, Romania). For preliminary molecular identification GenBank was searched for similar ITS sequences using blastn (Altschul et al. 1990).
However, according to Weir et al. (2012) Colletotrichum species could not reliably distinguished by using ITS sequence alone. Following their suggestions, the glyceraldehyde-3-phosphate dehydrogenase (GAPDH)-sequence was additionally used for species identification in this study. GAPDH was amplified using primers GDF/GDR (Templeton et al. 1992). The PCR-mix was set up in a reaction volume of 25 µl containing 1 u BioTherm Taq DNA polymerase (GeneCraft UK Products Semiramis Genetics Ltd., Manchester, UK), the corresponding 1 × buffer, 0.4 µM of each primer, 0.4 µM dNTP-mix, 1 µl Bovine Serum Albumin (BSA; 50 mg/ml), 1 µl genomic DNA (1:10 diluted). Thermocycling conditions were as follows: 4 min at 95 °C, 35 cycles at 95 °C for 30 s, 60 °C for 30 s, 72 °C for 45 s and a final extension at 72 °C for 7 min. The product was purified, Sanger sequenced and edited as written above. Both consensus sequences were deposited in GenBank (https://www.ncbi.nlm.nih.gov/Genbank/) under the accession numbers OP316868 (ITS) and OP345225 (GAPDH).
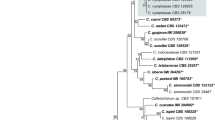
Detailed species identification was performed by creating a multi locus dataset of ITS and GAPDH reference sequences available in GenBank according to Liu et al. (2014), Crous et al. (2015) and Kee et al. (2020c). Phylogenetic analysis was performed by using MEGA X (Kumar et al. 2018). The sequences were aligned by using the incorporated software MUSCLE (Edgar 2004). Maximum likelihood (ML) interference was done using the Kimura 2‑parameter model (Kimura 1980) for substitution and 1000 bootstrap replicates. The tree was rooted against C. cliviae as an outgroup species.
Results
Microbiological Identification
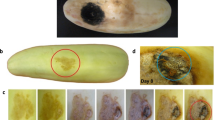
Visual inspection revealed on both sides of the leaves randomly distributed, initially small, roundish, water-soaked lesions, which rapidly enlarged and coalesced, eventually leading to severe leaf blight, sharply demarcated from healthy tissue. Concentric rings of dark acervuli sunken into the epidermis appeared on the dried lesions (Figs. 1 and 2).
Under humid conditions, the fruiting bodies produced a bright orange mass of elongated, straight to slightly irregular, cylindrical to slightly club-shaped conidia, obtuse to rounded at apex, truncated at base with acuminate attachment point (Fig. 3). The microscopic measurements of conidia length showed (12.3–) 18.7 ± 2.3 (–26.7) µm and a median of 18.4 µm. The width of the conidia was (3.3–) 6.1 ± 1.0 (–8.7) µm, the median 6.1 µm.
Measurements of appressoria (n = 50) revealed (6.57–) 9.00 ± 1.14 (–12.21) µm, median 8.84 µm.
After 14 days at 25 °C on PDA50% colonies measured (53.0–) 60.8 ± 4.6 (–68.0) mm, median 59.5 mm in diameter. They appeared white at the edge, grayish-white to partly cream to greyish purple and dark grey in the center, felted with aerial mycelium (Fig. 4).
Molecular Phylogeny
Based on BLAST search the isolate showed 98.8–99.8% identity with available C. sansevieriae sequences for ITS gene. In the Maximum Likelihood analysis of the combined dataset containing 460 sites, the isolate clustered within C. sansevieriae with high bootstrap support (99%) (Fig. 5).
Maximum likelihood tree inferred from combined ITS and GAPDH sequences of Colletotrichum species. The species name is followed by the strain number and GenBank accession numbers. The tree is rooted to C. cliviae. Bootstrap values (≥ 50%) are indicated at the nodes. The tree with the highest log likelihood (−1205.61) is shown
Discussion
Both the microbiological and the phylogenetic examination resulted in the conclusive detection of Colletotrichum sansevieriae as causal agent of the observed leaf spots. Macroscopic as well as morphological characteristics are in good accordance with data in literature (Nakamura et al. 2006; Park et al. 2013; Kee et al. 2020c). However, the appressoria measurements of this study are slightly larger than documented in the first description of the species (Nakamura et al. 2006), but agree well with those of Park et al. (2013) and Kee et al. (2020c). The differences may be due to different cultivation methods for appressoria production. This is the first report of the occurrence of this pathogen on Sansevieria trifasciata in Germany and more widely in Europe.
On many plant hosts, Colletotrichum belong to the most serious pathogens. Some Colletotrichum species have a very wide host range, while others are host-specific (Bhunjun et al. 2021; Talhinhas and Baroncelli 2021). On Sansevieria only two species are known to occur, C. sansevieriae and C. neosansevieriae. However, the latter being found only once in South Africa and seems to be extremely rare (Crous et al. 2015; Talhinhas and Baroncelli 2021). In contrast, C. sansevieriae is reported from several countries worldwide (Nakamura et al. 2006; Aldaoud et al. 2011; Campoverde and Palmateer 2012; Palmateer et al. 2012; Pérez-León et al. 2013; Gautam et al. 2012; Park et al. 2013; Karimi et al. 2017; Li et al. 2020; Kee et al. 2020b). So far known, the host plant range of C. sansevieriae is very narrow. All the above observations of natural infection were made on S. trifasciata or its cultivars. After artificial inoculation, besides S. trifasciata and its cultivars, only S. stuckyi (Nakamura et al. 2006) developed symptoms. Other Sansevieria species—S. cylindrica, S. masoniana—as well as other tested plants from various genera were not susceptible (Nakamura et al. 2006; Pérez-León et al. 2013; Kee et al. 2020c).
According to currently available data, the high host specificity of C. sansevieriae is outstanding compared to other Colletotrichum species (Nakamura et al. 2006; Kee et al. 2020c, Talhinhas and Baroncelli 2021). However, the number of tested potential host species remains very low and does not include close relatives of S. trifasciata (van Kleinwee et al. 2022). Further investigation with respect to the host range is needed to better understand the host specificity of C. sansevieriae.
Infection always required injury to the leaves; unwounded leaves could not be infected, although penetration into stomata was observed (Kee et al. 2020c). It appears that C. sansevieriae is a comparatively weak pathogen and the risks for Sansevieria producing nurseries are manageable if good professional practice and hygiene are observed. These include avoiding injury, keeping leaves dry, and eliminating diseased leaves or plants (Campoverde and Palmateer 2012). According to the same authors, treatments with modern fungicides (e.g., triazoles, strobilurins) are effective when applied prophylactically, but not curatively.
As the infected plants were imported from Costa Rica, where the occurrence of C. sansevieriae is known from farms producing Sansevieria for the markets in the USA and Europe (Pérez-León et al. 2013), the source of introduction is obvious. Apparently, the observation confirms the general risk of distributing plant pests and diseases by global plant trade (Hantula et al. 2014; Spence et al. 2020). However, at least for European horticulture, the occurrence of C. sansevieriae does not pose a significant risk: because of its high host specificity, low virulence, low economic importance of Sansevieria, its exclusive use as a houseplant, and easy-to-implement countermeasures. Serious economic as well as ecological damage is not to be expected.
References
Adeniyi AG, Akorede AS, Ighalo JO (2020) Sansevieria trifasciata fibre and composites: a review of recent developments. Int Polym Process 35:344–354. https://doi.org/10.3139/217.391
Ahmadpour A, Poursafar A (2018) Stemphylium vesicarium causing Sansevieria trifasciata (viper’s bowstring hemp) leaf blight in Iran. Australas Plant Dis Notes 13:4. https://doi.org/10.1007/s13314-018-0288-3
Aldaoud R, DeAlwis S, Salib S, Cunnington JH, Doughty S (2011) First record of Colletotrichum sansevieriae on Sansevieria sp. (mother-in-law’s tongue) in Australia. Australas Plant Dis Notes 6:60–61. https://doi.org/10.1007/s13314-011-0020-z
Altschul SF, Gish W, Miller W, Myers EW, Myers DJ, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215:403–410
Bhunjun CS, Phukhamsakda C, Jayawardena RS, Jeewons R, Promputtha I, Hyde KD (2021) Investigating species boundaries in Colletotrichum. Fungal Divers 107:107–127. https://doi.org/10.1007/s13225-021-00471-z
Brink M, Achigan-Dako EG (eds) (2012) Plant resources of tropical africa 16. fibres. PROTA Foundation, Wageningen
Campoverde EV, Palmateer AJ (2012) Colletotrichum sansevieriae causing anthracnose of Sansevieria trifasciata ‘Laurentii’ and ‘Moonshine’ in South Florida. Proc Fla State Hortic Soc 125:359–360
Crous PW, Wingfield MJ, Guarro J, Hernández-Restrepo M, Sutton DA et al (2015) Fungal Planet description sheets: 320–370. Persoonia 34:167–266
Edgar RC (2004) MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Research 32(5):1792-1797. https://doi.org/10.1093/nar/gkh340
Fiscal RR, Dandan KBV (2016) Development and evaluation of paper from Corn husks (Zea mays L.) and snake plant fibers (Sansevieria zeylanica). IJSR 5:867–870. https://doi.org/10.21275/v5i8.3081601
Gardes M, Bruns TD (1993) ITS primers with enhanced specificity for basidiomycetes—application to the identification of mycorrhizae and rusts. Mol Ecol 2:113–118. https://doi.org/10.1111/j.1365-294X.1993.tb00005.x
Gautam AK, Avashti S, Bhadauria R (2012) Colletotrichum sansevieriae on Sansevieria trisfasciata—a report from Madhya Pradesh, India. Plant Pathol Quar 2:190–192. https://doi.org/10.5943/ppq/2/2/12
Hantula J, Müller M, Uusivuori J (2014) International plant trade associated risks: Laissez-faire or novel solutions. Environ Sci Policy 37:158–160
Henley RW (1982) Sansevieria in Florida—past and present. Proc Fla State Hortic Soc 95:295–298
Karimi O, Tajick Ghanbari MA, Bagherabadi S, Moradi Amirabad Y (2017) First report of Colletotrichum sansevieriae causing anthracnose on Sansevieria trifasciata. Iran J Plant Pathol 99:302
Kee YJ, Zakaria L, Mohd MH (2017a) First report of Stemphylium lycopersici causing leaf spot on Sansevieria trifasciata in Malaysia. Plant Dis 102:445–446. https://doi.org/10.1094/PDIS-07-17-1049-PDN
Kee YJ, Suhaimi NN, Zakaria L, Mohd MH (2017b) Characterisation of Neoscytalidium dimidiatum causing leaf blight on Sansevieria trifasciata in Malaysia. Australas Plant Dis Notes 12:60
Kee YJ, Zakaria L, Mohd MH (2019) Lasiodiplodia species associated with Sansevieria trifasciata leaf blight in Malaysia. J Gen Plant Pathol 85:66–71. https://doi.org/10.1007/s10327-018-0814-3
Kee YJ, Zakaria L, Mohd MH (2020a) Curvularia asianensis and Curvularia eragrostidis associated with leaf spot of Sansevieria trifasciata in Malaysia. J Phytopathol 168:290–296. https://doi.org/10.1111/jph.12890
Kee YJ, Zakaria L, Mohd MH (2020b) Morphology, phylogeny and pathogenicity of Fusarium species from Sansevieria trifasciata in Malaysia. Plant Pathol 69:442–454. https://doi.org/10.1111/ppa.13138
Kee YJ, Zakaria L, Mohd MH (2020c) Identification, pathogenicity and histopathology of Colletotrichum sansevieriae causing anthracnose on Sansevieria trifasciata in Malaysia. J Appl Microbiol 129:626–636
Kimura M (1980) A simple method for estimating evolutionary rate of base substitutions through comparative studies of nucleotide sequences. J Mol Evol 16:111–120
van Kleinwee I, Larridon I, Shah T, Bauters K, Asselman P, Goetghebeur P, Leliaert F, Veltjen E (2022) Plastid phylogenomics of the Sansevieria Clade of Dracaena (Asparagaceae) resolves a recent radiation. Mol Phylogenet Evol 169:107404. https://doi.org/10.1016/j.ympev.2022.107404
Kumar S, Stecher G, Li M, Knyaz C, Tamura K (2018) MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol 35(6):1547-1549. https://doi.org/10.1093/molbev/msy096
Li W‑J, McKenzie EHC, Liu J‑K, Bhat DJ, Dai D‑Q, Camporesi E, Tian Q, Maharachchikumbura SSN, Luo Z‑L, Shang Q‑J, Zhang J‑F, Tangthirasunun N, Karunarathna SC, Xu J‑C, Hyde KD (2020) Taxonomy and phylogeny of hyaline-spored coelomycetes. Fungal Divers 100:279–801. https://doi.org/10.1007/s13225-020-00440-y
Li YL, Zhou Z, Lu W, Ye JR (2013) First report of a Chaetomella sp. causing a leaf spot on Sansevieria trifasciata in China. Plant Dis 97:992. https://doi.org/10.1094/PDIS-10-12-0993-PDN
Liu F, Cai L, Crous PW, Damm U (2014) The Colletotrichum gigasporum species complex. Pers Mol Phylogeny Evol Fungi 33:83–97. https://doi.org/10.3767/003158514X684447
Mohana VR, Rajeshb A, Athiperumalsamia T, Sutha S (2008) Ethnomedicinal plants of the Tirunelveli District, Tamil Nadu, India. Ethnobot Leafl 12:79–95
Monteles RP, Sousa ES, da Matos SK, de Brito VST, de Melo MP, Beserra JEA (2020) Neoscytalidium dimidiatum causes leaf blight on Sansevieria trifasciata in Brazil. Australas Plant Dis Notes 15:19. https://doi.org/10.1007/s13314-020-00389-6
Nakamura M, Ohzono M, Iwai H, Arai K (2006) Anthracnose of Sansevieria trifasciata caused by Colletotrichum sansevieriae sp. nov. J Gen Plant Pathol 72:253–256
Nakamura M, Fujikawa T, Nakamori D, Iwai H (2018) Draft genome sequence of Colletotrichum sansevieriae Sa-1‑2, the anthracnose pathogen of Sansevieria trifasciata. Data Brief 18:691–695. https://doi.org/10.1016/j.dib.2018.03.083
Newton LE (2020) Sansevieria ruscaceae. In: Eggli U, Nyfeller R (eds) Bromeliaceae to xanthorrhoeaceae, 2nd edn. Illustrated handbook of succulent plants—Monocotyledons, vol 2. Springer, Berlin, pp 271–284
Palmateer AJ, Tarnowski TLB, Lopez P (2012) First report of Colletotrichum sansevieriae causing anthracnose on Sansevieria trifasciata in Florida. Plant Dis 96:293. https://doi.org/10.1094/pdis-06-11-0539
Park JH, Han KS, Kim JY, Shin HD (2013) First report of anthracnose caused by Colletotrichum sansevieriae on Sansevieria in Korea. Plant Dis 97:1510. https://doi.org/10.1094/PDIS-04-13-0402-PDN
Pérez-León G, Chavarría-Pérez L, Araya-Quesada J, Gómez-Alpízar L (2013) Identificación del agente causal de la antracnosis de Sansevieria spp. en Costa Rica. Agron Costarric 37:39–50
Smith BJ, Black LL (1990) Morphological, cultural, and pathogenic variation among Colletotrichum species isolated from strawberry. Plant Dis 74:69–76
Spence N, Hill L, Morris J (2020) How the global threat of pests and diseases impacts plants, people, and the planet. Plants People Planet 2:5–13
Takawira-Nyenya R, Stedje B (2011) Ethnobotanical studies in the genus Sansevieria Thunb. (Asparagaceae) in Zimbabwe. Ethnobot Res Appl 9:421–444. https://doi.org/10.17348/era.9.0.421-443
Takawira-Nyenya R, Newton LE, Wabuyele E, Stedje B (2014) Ethnobotanical uses of Sansevieria Thunb (Asparagaceae) in coast province of Kenya. Ethnobot Res Appl 12:51–69
Takawira-Nyenya R, Mucina L, Cardinal-McTeague WM, Thiele K (2018) Sansevieria (Asparagaceae, Nolinoideae) is a herbaceous clade within Dracaena: inference from non-coding plastid and nuclear DNA sequence data. Phytotaxa 376:254. https://doi.org/10.11646/phytotaxa.376.6.2
Talhinhas P, Baroncelli P (2021) Colletotrichum species and complexes: geographic distribution, host range and conservation status. Fungal Divers 110:109–198. https://doi.org/10.1007/s13225-021-00491-9
Templeton MD, Rikkerink EHA, Solon SL, Crowhurst RN (1992) Cloning and molecular characterization of the glyceraldehyde-3-phosphate dehydrogenase encoding gene and cDNA from the plant pathogenic fungus Glomerella cingulata. Gene 122:225–230
Weir BS, Johnston PR, Damm U (2012) The Colletotrichum gloeosporioides species complex. Stud Mycol 73(1):115–180. https://doi.org/10.3114/sim0011
White TJ, Bruns T, Lee S, Taylor JW (1990) Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, Sninsky JJ, White TJ (eds) PCR protocols: A guide to methods and applications. Academic Press, New York, pp 315–322
Acknowledgements
We are grateful to Lucija Jahn, Merle Bröring, and Jenny Rebentisch for their dedicated technical support.
Funding
Open Access funding enabled and organized by Projekt DEAL.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
T. Brand and A. Wichura declare that they have no competing interests.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Brand, T., Wichura, A. First Report on Colletotrichum sansevieriae Causing Anthracnose of Sansevieria trifasciata in Germany. Gesunde Pflanzen 75, 61–66 (2023). https://doi.org/10.1007/s10343-022-00777-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10343-022-00777-1