Abstract
RNA interference (RNAi) regulates gene expression in eukaryotes, and it is an emerging tool in crop protection by exogenous applications of double-stranded RNAs (dsRNAs) to silence the expression of essential pest genes. Nevertheless, delivery of dsRNAs to sap-sucking insects is a major challenge for RNAi applications. The present work aimed at verifying whether in leafhopper species, RNAi can be triggered by plant-mediated delivery, and providing a proof of concept towards field applications. Two phytoplasma vectors species, Euscelidius variegatus and Scaphoideus titanus (Hemiptera: Cicadellidae), were used as case study. Gene silencing can be achieved efficiently in both species through microinjection of dsRNAs, despite the technique being time consuming and inapplicable on large scale. This protocol was set as gold standard for the development of a higher throughput approach. Soaking of nymphs in a solution with co-adjuvant and dsRNAs as well as insect feeding on whole plants or detached leaves immersed in a dsRNA solution were assayed as alternative delivery strategies. Nymph soaking did not induce specific gene silencing, while plant absorption proved to be suitable to deliver both a coloured solution and control dsRNAs targeting green fluorescent protein gene. Insect feeding on detached leaves immersed in dsRNA solution was selected to test silencing of two gut-specific (legumain and natterin) and one ubiquitous (ATP synthase β) genes. The expression of the three genes significantly decreased in E. variegatus insects fed on dsRNA-treated plants. Similarly, a significant reduction of ATP synthase β transcript was measured in S. titanus fed on dsRNA-treated plants.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Key message
-
Application of RNAi as bio-insecticide against phloem-feeders is hampered by dsRNA delivery issues
-
RNAi occurs in leafhopper phytoplasma vectors Scaphoideus titanus and Euscelidius variegatus
-
Dye-solution, adsorbed by plants, moves to xylem and stains guts of insects fed on treated plants
-
Three target genes were silenced in insects fed on detached plant leaves that absorbed dsRNAs
-
Soaking of E. variegatus nymphs in dsRNAs + lipofectamin did not induce specific gene silencing
Introduction
RNA interference (RNAi) is a sequence-specific mechanism of eukaryotes (Fire et al. 1998; Baulcombe 2004) regulating gene expression and providing the primary natural defence against nucleic acids of transposons or viruses (Santos et al. 2019; Bonning and Saleh 2021). Strategies based on RNAi were originally exploited as molecular tools for studying gene function, and are now emerging in crop protection as powerful and precise techniques to develop new control tactics against plant pathogenic fungi, viruses and insect pests (Mezzetti et al. 2020; Christiaens et al. 2022; Wei et al. 2023). The technology is based on the use of double-stranded RNAs (dsRNAs) to silence the expression of essential genes of a selected pest. Systemic RNAi is well-documented for plants, fungi and nematodes, in which RNA-dependent RNA polymerases (RdRPs) amplify the triggering dsRNAs by generating secondary siRNAs that spread within the organism (Pinzón et al. 2019; Liu et al. 2020). The basic mechanism is known in arthropods, but knowledge gaps still exist. In fact, RdRPs have not been found in insects (Tomoyasu et al. 2008; Zong et al. 2009; Li et al. 2018; Christiaens et al. 2020b) and mechanisms of systemic RNAi are yet to be deciphered (Huvenne and Smagghe 2010; Vélez and Fishilevich 2018). Another unsolved question is the variable efficiency of RNAi mechanisms among insects (Silver et al. 2021). Species of the order Coleoptera display very robust RNAi responses (Baum et al. 2007; Zhu et al. 2011; Palli 2014), whereas lepidopteran and dipteran species respond to naked dsRNAs through injection but only in some cell types (Miller et al. 2008; Whyard et al. 2009; Terenius et al. 2011; Zhu and Palli 2020). Several constraints still hamper the open-field application of RNAi in agriculture, such as the non-harmonized legislation regulating the use of exogenous dsRNAs, which has so far been approved in some non-EU countries (OECD 2020; Mezzetti et al. 2020; Dietz-Pfeilstetter et al. 2021), the cost-effective production of dsRNAs (Palli 2014), the lack of risk assessment protocols for undesirable off-target effects (Papadopoulou et al. 2020), possible instability of dsRNA molecules to atmospheric agents (Yu et al. 2013; Christiaens et al. 2020b), delivery issues of dsRNAs, especially in the case of sap-sucking insects (Christiaens and Smagghe 2014; Coleman et al. 2015; Yu et al. 2016). Focusing on the latter issue, it is worth mentioning that, unlike chewing insects, delivery of dsRNAs to sap-sucking insects is a major challenge for RNAi application in crop protection, and only few treatment strategies, mainly based on nanoengineered formulations, have been proposed (Yu et al. 2013; Ghosh et al. 2017; Christiaens et al. 2020a; Pugsley et al. 2021; Jain et al. 2022).
According to previous results (Dalakouras et al. 2018), dsRNAs applied directly into plant vascular tissues using trunk injection and petiole absorption are taken up and systemically transported, but they remain restricted to the xylem and the apoplast. This aspect might be particularly promising for sap-sucking species since electro-penetrography data indicate that phloem-sucking leafhoppers (such as the big majority of phytoplasma vectors) also feed on the xylem (Lett et al. 2001; Stafford and Walker 2009; Carpane et al. 2011; Trębicki et al. 2012; Chuche et al. 2017; Ripamonti et al. 2022b). Indeed, phloem-, xylem- and parenchyma feeding behaviours (Tonkyn and Whitcomb 1987) are not strict categories (Wayadande 1994).
The aim of this research is to verify whether the exogenous application of dsRNAs directly to the plant vascular system may trigger RNAi in two leafhopper species, Scaphoideus titanus and Euscelidus variegatus. These species are the vectors of the quarantine pest Flavescence dorée phytoplasma (FDp) (EFSA (European Food Safety Authority) et al. 2019), enquired in question N°, in natural and laboratory conditions, respectively (Caudwell et al. 1972; Chuche and Thiéry 2014). Phytoplasmas are obligate intracellular bacteria colonizing the plant phloem and several organs of their insect vectors and are responsible for numerous plant diseases. Few complete phytoplasma genomes and several partial drafts highlight the reduced genome size and lack of crucial metabolic pathways (e.g. ATP synthesis, de novo synthesis of nucleotides), which may result from their endo-parasitic lifestyle (Oshima et al. 2013; Debonneville et al. 2022). Grapevine Flavescence dorée is a devastating disease that is widespread in the vast majority of grapevine-growing areas of Europe (EFSA Panel on Plant Health PLH 2014; Tramontini et al. 2020). The persistent propagative transmission is driven by molecular interactions between vector proteins and FDp membrane proteins (Arricau-Bouvery et al. 2018, 2021, 2023; Trivellone et al. 2019), and specific phytoplasma proteins are associated with epidemic FDp strains transmitted by S. titanus (Malembic-Maher et al. 2020). Flavescence dorée management relies conventionally on control of vector population through insecticide applications, rogueing of infected plants to reduce inoculum loads and planting of phytoplasma-free grafted cuttings (Oliveira et al. 2019). The RNAi mechanism efficiently occurs in both S. titanus and E. variegatus following injection of dsRNAs (Abbà et al. 2019; Ripamonti et al. 2022a), but this delivery strategy is manageable only under lab conditions and is poorly suitable to treat nymphal stages. Delivery methods other than dsRNA-injection, that can be more suitable for possible future field applications, were attempted in this work. Soaking of nymphs in a solution with co-adjuvant and dsRNAs as well as insect feeding on detached leaves immersed in a dsRNA solution were assayed as alternative delivery strategies. Two gut-specific and one ubiquitous transcripts were selected as target genes to prove the effectiveness of gene silencing in insects fed on dsRNA-treated plants.
Materials and methods
Insect rearing and plant production
Euscelidius variegatus was originally collected in Piedmont and continuously reared on oat (Avena sativa (L.)) plants from seed, inside plastic and nylon cages in growth chambers at 20–25 °C with a L16:D8 photoperiod. Scaphoideus titanus has one generation per year and its continuous rearing under controlled conditions is not feasible. To obtain coeval S. titanus specimens, two-year-old grapevine canes bearing leafhopper eggs were collected in Veneto (Italy) vineyards during the winter period and kept at 5 ± 1 °C. To allow coordinated egg hatching, grapevine branches were caged inside insect-proof screen houses in a glasshouse with natural light and temperature ranging from 20 to 25 °C. Potted grapevine (Vitis vinifera L.) cuttings and healthy broad bean (Vicia faba L.) plants from seed were introduced in the screen house to feed the newly hatched nymphs. Third to fifth instar nymphs were used for dsRNA treatment by soaking (E. variegatus) and feeding (E. variegatus and S. titanus), whereas coeval newly emerged E. variegatus adults were used for injection of designed dsRNAs (targeting legumain and natterin genes) to assess their silencing efficiency.
Synthesis of dsRNAs
The complete coding sequences of E. variegatus genes, ATP synthase β, legumain and natterin selected to be silenced by RNAi were found in the TSA sequence database (BioProject: PRJNA393620) at NCBI under the accession numbers GFTU01013594.1, GFTU01008326.1 and GFTU01002968.1, respectively. ATP synthase β was selected as systemically expressed and involved in phytoplasma transmission (Galetto et al. 2011, 2021), whereas the other two (legumain and natterin) are expressed in gut tissue (Hartmann et al. 2018; Molina-Cruz et al. 2020), the first site where dsRNAs acquired by feeding may activate RNAi (Kunte et al. 2020). The complete coding sequence of S. titanus ATP synthase β (GenBank accession number: MZ130944) was retrieved from a transcriptome project aimed at describing virus (Ottati et al. 2020) and microbial populations (Abbà et al. 2022) of this insect species.
Fragments of the target sequences were obtained from total RNA isolated from adult insects using a reverse transcription polymerase chain reaction (RT-PCR). A control template corresponding to a fragment of the gene sequence of green fluorescent protein (GFP) was PCR-amplified from plasmid pJL24 (Lindbo 2007). Primers used to generate the dsRNA templates included the T7 promoter sequence at their 5’-end (Table S1). The PCR products were ligated into the pGEM-T Easy plasmid (Promega) and the plasmids were used as templates for the subsequent PCRs. Then, 1 μg of each column-purified PCR product was in vitro transcribed using the MEGAscript RNAi Kit (Thermo Fisher Scientific) according to the manufacturer’s instructions. After column-purification with ssDNA/RNA Clean and Concentrator (Zymo Research) and elution in Tris–EDTA buffer (10 mM Tris–HCl, 0.1 mM EDTA, pH 8.5), dsRNAs were quantified using a Nanodrop ND-1000 spectrophotometer (Thermo Fisher Scientific).
Delivery strategies
Insect soaking
About 20 E. variegatus second-instar nymphs were submerged in a watch glass with 200 μl soaking solution containing dsRNAs (1μγ/μl), targeting either E. variegatus ATP synthase β gene or GFP, used as negative control. The soaking solution contained lipofectamine (Thermo Fisher Scientific, USA) at a final concentration of 13%, to help internalization of dsRNAs molecules. Nymphs were soaked at room temperature for 5 min. Then, nymphs were dried on paper towels and then kept for 8 d on A. sativa before RNA extraction and gene expression analysis by qPCR. The experiment was repeated three times.
Petiole absorption
The method was optimized on V. faba and V. vinifera whole plants using a solution of diluted blue food colouring (Fig. S1). The first basal leaf was detached from 12 cm tall broad beans and from 20 cm tall micropropagated grapevines and the remaining protruding petioles were attached to 1.5 ml tubes with cut off bottom and containing 400 μL of diluted food colour solution (Fig. S1). After 2 h the apical leaves were observed to trace the movement of the colour solution.
In order to assess if a solution containing dsRNAs absorbed by plants could be acquired by insects, ten E. variegatus adults were fed for 48 h on treated broad beans: bottomless 1.5 ml tubes containing 400 μL of dsGFP solution (7 μg/tube) were attached to the protruding petioles of basal leaves, as described above. Insects were then isolated on oat for eight days and collected. Total RNA was extracted from single specimens and subjected to retro-transcription and PCR amplification with GFP specific primers (Table S1), as detailed below.
Detached leaf absorption
Single plant leaves instead of whole plants were allowed to absorb a solution of diluted blue food colouring (Fig. 1), in order to concentrate it on a smaller plant surface. Single leaves detached from V. faba, V. vinifera and A. sativa were partly inserted in 1.5 ml tubes containing the colour solution and partly kept out from the solution. After 2 h, the upper leaf parts (not immersed in the solution) were analysed under the microscope to trace the movement of the colour marker. Free-hand sections from treated plant leaves of V. faba, V. vinifera and A. sativa were observed under a Leica DM 750 microscope equipped with a EC4 camera. Microscope images were then processed with the ImageJ (1.46r, NIH, https://imagej.nih.gov) software.
Absorption of a diluted food colouring solution by broad bean (left), grapevine (middle) and oat (right) detached leaves a Microscopic observations of marker diffusion in free-hand sections of treated broad bean b, grapevine c and oat d leaves. Black arrows indicate blue stained tissues. Bars 200 μm
In order to assess if solution absorbed by plants could be acquired by insects, E. variegatus adults were fed for 48 h on blue-treated leaves of A. sativa (Fig. 2). Insects were then anaesthetized with CO2 and guts were dissected with forceps using a stereomicroscope (Leica S9i) in a drop of phosphate-buffered saline (PBS), mounted on glass slides and observed with an optic microscope, to detect the entrance of the colour solution in insect body. A scale of blue 0–3 (index 0 = no staining, index 1 = weak staining, index 2 = average staining, index 3 = strong staining) was set up to record the intensity of gut staining. Dissected E. variegatus guts were observed under a Leica DM 750 microscope and processed as described above.
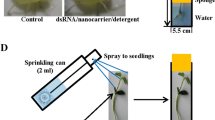
Absorption by detached oat and grapevine leaves was then chosen as final plant-mediated strategy to deliver dsRNAs to insects. Detached leaves were inserted in 1.5-ml tubes containing 200 μL of dsRNA solution (7 μg/tube), until the solution was all absorbed (roughly 2 h); 1 ml of tap water was then added to each tube and tubes were closed with parafilm to prevent evaporation. The dsRNA-treated leaves were used to feed E. variegatus (about 20 fifth-instar nymphs on each oat leaf) and S. titanus fifth-instar nymphs (about 20 on each grapevine leaf), for 48 h. Insects were then transferred on fresh oat (E. variegatus) or grapevine (S. titanus) plants, prior to RNA extraction and gene expression analysis.
Abdominal microinjection
Microinjection of dsRNAs has been optimized and proven to work well as delivery strategy in both E. variegatus and S. titanus in previous works (Abbà et al. 2019; Ripamonti et al. 2022a). Newly emerged adults were anaesthetized with CO2 and microinjected between two abdominal segments under a stereomicroscope using a fine glass needle connected to a Cell Tram Oil microinjector (Eppendorf, Hamburg, Germany). Insects were microinjected with 0.5 μL of dsRNAs at the concentrations of 160 ng/μL (80 ng of dsRNA/insect). Injected insects were then caged on oat plants and monitored daily until the end of the experiment (seven days post-injection). This delivery strategy was used here to determine silencing efficiency of the newly designed dsRNAs (targeting E. variegatus legumain and natterin genes). Silencing efficiency of dsRNAs targeting ATP synthase β of both species has been previously assessed by microinjection (Abbà et al. 2019; Ripamonti et al. 2022a).
RNA extraction
Total RNAs were extracted from single insect specimen, in order to determine insect acquisition of GFP-dsRNAs after petiole absorption (E. variegatus) as well as to evaluate the reduction of specific target transcripts in dsRNA-treated insects (E. variegatus or S. titanus). The samples were frozen in liquid nitrogen, crushed with a micropestle in sterile Eppendorf tubes, and homogenized in 0.5 ml Tri-Reagent (Zymo Research). Samples were centrifuged 1 min at 12.000 g at 4 °C and RNAs were extracted from supernatants with Direct-zol RNA Mini Prep kit (Zymo Research), following manufacturer’s protocol and including the optional DNAse treatment step. Concentration, purity, and quality of extracted RNA samples were analysed in a Nanodrop ND-1000 spectrophotometer (Thermo Fisher Scientific).
Total RNAs were extracted from dissected organs from E. variegatus, in order to evaluate the tissue-specific expression of legumain and natterin transcripts. In particular, RNAs were extracted from salivary glands, guts, ovaries and testes of newly emerged adults. For each organ, four pooled samples (each made of ten dissected organs) were obtained. Organs were carefully isolated from CO2 anaesthetized insects with forceps and needles using a stereomicroscope, rinsed with phosphate-buffered saline (PBS) solution and immediately frozen in liquid nitrogen. Total RNAs were extracted as described above.
RT-PCR and gene expression studies
Qualitative RT-PCR was used to detect dsGFPs in insects fed on V. faba plants after petiole absorption. For each sample, cDNA was synthesized from 300 ng of total RNA with random hexamers using a High Capacity cDNA reverse transcription kit (Applied Biosystems). The resulting cDNA (1 μL) was used as a template for qPCR in a 20 μL volume mix, containing Taq DNA polymerase (1 U) (Polymed) and 350 nM of each primer. The GFP specific primer pairs used for the PCR are listed in Table S1. The cycling conditions were set as follow: 2 min at 94 °C and 35 cycles with 1 cycle consisting of 30 s at 94 °C, 30 s at 62 °C and 40 s at 72 °C followed by a final extension of 5 min at 72 °C. PCR products were analysed by electrophoresis through 1% agarose gel in 1 × Tris–borate-EDTA (TBE) buffer, along with a 1-kb-plus DNA size marker (Gibco BRL). Gels were stained with ethidium bromide and visualized on a UV transilluminator.
Quantitative RT-PCR was used to quantify the ability of the administered dsRNAs to knockdown target mRNAs (ATP synthase β, legumain and natterin), as well as to determine tissue specific expression of E. variegatus legumain and natterin transcripts. A minimum of eight to a maximum of 48 insect biological replicates were analysed for each treated group in the different described experiments, as detailed in Table 1 and in Supplementary Tables S2–S5. For each sample, cDNA was synthesized from total RNA (500 ng from whole insect samples and 150 ng from dissected organ samples) with random hexamers using a High Capacity cDNA reverse transcription kit (Applied Biosystems). The resulting cDNA (1 μL) was used as a template for qPCR in a 10 μL volume mix, containing 1 × iTaq Universal Sybr Green Supermix (Bio-Rad) and 300 nM of each primer. All the primer pairs used for qPCR are listed in Table S1. Samples were run in duplicate in a CFX Connect Real-Time PCR Detection System (Bio-Rad). Cycling conditions were: 95 °C for 3 min, and 40 cycles at 95 °C for 15 s and 60 °C for 30 s of annealing/extension step. The specificity of the PCR products was verified by melting curve analysis for all samples. No-template controls were always included in each plate. Primers targeting glutathione S-transferase and elongation factor-1α (Galetto et al. 2013, 2018; Ripamonti et al. 2022a) were used as internal controls to normalize the cDNA among samples. Normalized expression levels of each target gene for each sample were calculated by CFXMaestro™ Software (Bio-Rad). The expression stability of reference genes was acceptable in the multiplate gene study.
Statistical analysis
dsRNA-treated insects with expression level higher than mean and median of the corresponding dsGFP-treated groups were excluded from the statistical analysis (Supplementary Tables S4 and S5); according to the interquartile range (IQR) method, these excluded samples were indeed outliers (1.5 × IQR) or extreme outliers (3 × IQR). When raw transcript data were not normally distributed, they were natural log-transformed before analysis. ANOVA was used to compare transcript levels measured in different E. variegatus dissected organs. Parametric t test or nonparametric Mann–Whitney test ( when data were not normally distributed) were used to compare levels of different transcripts measured in E. variegatus adults injected with dsRNAs as well as in E. variegatus and in S. titanus insects fed on dsRNA-treated plants (dsGFP vs. dsRNAs targeting ATP synthase β, legumain or natterin genes). SigmaPlot version 13 (Systat Software, Inc., San Jose, CA, USA) was used for statistical analyses.
Results
Optimization of delivery strategies
All the experiments to set up delivery methods were initially performed on E. variegatus, in order to apply the most performing protocol in a further step to the economically important association S. titanus/grapevine.
Insect soaking
The delivery of dsRNA through soaking was attempted on E. variegatus nymphs with dsRNAs targeting ATP synthase β, using lipofectamine as adjuvant of the soaking treatment as preliminary microscopy observations indicated that this compound seemed to improve insect cuticle penetration (not shown). However, we could not detect the expected reduction of the specific target transcript compared with the expression level of the same gene measured in control samples (Table 1). Soaking was therefore excluded from further assessment on the S. titanus system.
Petiole absorption
Pilot experiments were conducted using a diluted blue food colouring solution absorbed by whole broad bean and grapevine plants. The blue solution applied to the protruding petioles of basal leaves moved to the apical leaves in about two hours, as shown by coloured leaf portions of both plant species (Fig. S1).
A preliminary assay to investigate the possibility of delivering dsRNAs to sap-feeding insects through the plant vascular system was conducted by treating whole V. faba plants with dsGFP and by feeding E. variegatus adults on treated broad beans. In two out of five insects dsGFPs were detected, even if the signal was faint, probably due to the high dilution of the dsRNA molecules in the whole plant tissues (data not shown). Therefore, we explored the possibility of using detached leaves (Fig. 1), instead of the entire plant, in order to deliver a higher concentration of dsRNAs to sap-feeding insects.
Detached leaf absorption
Pilot experiments were conducted using a diluted blue food colouring solution absorbed by detached broad bean, grapevine and oat leaves. The uptake and distal translocation of the blue solution through xylem vessels were visualized under the microscope (Fig. 1).
Due to the lowest dilution of the target molecules in the vascular system of its small leaves, oat was chosen for further optimization experiments aiming at increasing the insect probability of acquiring the target molecule. Adults of E. variegatus (n = 35), caged in a glass tube, were fed on detached oat leaves that had absorbed the food colouring solution to monitor the insect uptake of the blue (Fig. 2a). After 1 h, a paper disc placed on the bottom of the glass tube was blue-stained by insect honeydews (not shown). After 48 h, insect guts were dissected and visualized under a microscope. Overall, 71% of the observed guts were blue stained, and 26% of them showed high staining intensities (Fig. 2b–f).
Optimization of insect gene silencing by detached leaf absorption of dsRNAs
On the basis of the staining results, absorption by detached leaves was chosen as plant-mediated strategy to deliver dsRNAs to insects and was initially optimized on E. variegatus. Gut-specific transcripts were selected for optimization, as they are specifically expressed in the first organ that dsRNAs encounter after their acquisition via ingestion (Kunte et al. 2020).
Tissue-specific expression of gut-specific transcripts
Legumain and natterin genes are known to be expressed in gut tissue (Hartmann et al. 2018; Molina-Cruz et al. 2020) and were therefore selected as target genes to be silenced by plant-delivered dsRNAs. The transcript level of these genes was investigated in different E. variegatus tissues to confirm their specific expression in guts. Both legumain and natterin genes were more expressed in E. variegatus guts than in salivary glands, ovaries and testes (ANOVA: legumain F = 327.315, P < 0.001; natterin F: 151.338, P < 0.001) (Fig. 3, Supplementary Table S2). In particular, the legumain transcript was expressed averagely 2500, 139,000 and 8000 times more in gut tissues than in salivary glands, ovaries and testes, respectively. Natterin gene was expressed 260, 1500 and 18 times more in gut tissues than in salivary glands, ovaries and testes, respectively (Supplementary Table S2).
Silencing efficiency of dsRNAs targeting gut-specific transcripts
dsRNAs targeting legumain and natterin were then synthetized and, as preliminary step, their efficacy in triggering RNAi signal was assessed by microinjection, under conditions previously optimized for effective and robust gene silencing in E. variegatus (Abbà et al. 2019). The dsRNAs efficiently silenced their specific target transcripts following abdominal microinjection (Fig. 4, Supplementary Table S3). The normalized expression levels of legumain and natterin measured in insects injected with specific dsRNAs were significantly lower than those quantified in control dsGFP-treated specimens at seven days post-injection (t test: legumain t = − 11.135, P < 0.001; natterin t = − 14.244, P < 0.001). In particular, the reduction of specific transcripts ranged from 14 to 32-fold in insects injected with dsRNAs targeting legumain and natterin, respectively.
Over time silencing effects by dsRNAs acquired via detached leaf absorption
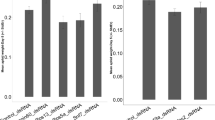
The silencing efficiency of dsRNAs targeting E. variegatus gut transcripts following plant-mediated delivery was assessed at 13, 17 and 24 days post-feeding. Euscelidius variegatus insects were fed on detached oat leaves previously treated with a solution containing dsRNAs (dsGFP/dsLeg/dsNat) and showed a significant reduction of the corresponding transcripts (legumain and natterin) at 17 and 24 days post-feeding (dpf), in comparison with insects fed on leaves treated with dsGFP (t test: legumain at 17 dpf t = − 1.969, P = 0.045 and at 24 dpf t = − 2.030, P = 0,032; natterin at 17 dpf t = − 2.005, P = 0.046 and at 24 dpf t = − 2.866, P = 0.006) (Fig. 5, Supplementary Table S4). Insects showing an expression level higher than the mean and the median values of dsGFP-treated samples were excluded from the statistical analysis as outliers. This was the case for three samples out of 18 insects treated with dsLeg (17%) and seven out of 21 treated with dsNat (33%) (Supplementary Table S4). In these outliers an insufficient or null uptake of dsRNAs might have occurred, as suggested by the preliminary experiments with the uptake of the blue solution, which failed to stain insect guts in 28% of the cases.
Expression level of Euscelidius variegatus legumain a and natterin b genes after dsRNA delivery through detached leaf absorption, measured at 13, 17 and 24 days post-feeding (dpf). dsGFP, dsLeg and dsNat indicate dsRNAs targeting green fluorescent protein, legumain and natterin genes, respectively. Asterisks indicate significant different expression levels; n.s.: not significant
Silencing of systemically expressed insect genes through plant-mediated dsRNA absorption
As a further step, the experimental conditions used to silence genes specifically expressed in guts were in the end applied to silence the systemically expressed ATP synthase β gene, involved in phytoplasma transmission (Galetto et al. 2011, 2021). Silencing of this gene was assessed in both E. variegatus and S. titanus insects, fed on detached leaves of oat (the former) and grapevine (the latter), after absorption of the dsRNA containing solution (Fig. 6, Supplementary Table S5). Specimens of both species showed a significant reduction of ATP synthase β transcript in comparison with insects fed on leaves treated with dsGFP (t test for E. variegatus: t = 7.351, P = < 0.001; Mann–Whitney test for S. titanus: U = 211.000, P = 0.002). Also, in this case, delivery of dsRNAs did not occur in all samples: two samples out of eight E. variegatus individuals (25%) and seven out of 25 S. titanus insects (28%) fed on dsATP-treated plants showed an expression level higher than the mean and the median values of corresponding dsGFP-treated samples, and were excluded from the statistical analysis as not-silenced (outliers, Supplementary Table S5). Even in this case the outliers might represent insects in which the dsRNA uptake did not occur efficiently.
Expression level of Euscelidius variegatus a and Scaphoideus titanus b ATP synthase β gene measured in insects sampled at 14 days post-feeding on dsRNA-treated detached oat and grapevine leaves, respectively. dsGFP and dsATP indicate dsRNAs targeting green fluorescent protein and ATP synthase β, respectively. Different letters indicate significant different expression levels
Discussion
RNA interference is a powerful tool to study gene function at the molecular level, but also an attractive tool for pest control, with a particular interest in the possible application of exogenous dsRNAs directly on crops. The key challenge for the practical use of RNAi as biopesticides for insect pest control is finding effective and reliable methods to deliver stable dsRNAs. Limited uptake and availability of the dsRNAs have restricted the development of RNAi-based biopesticides against sap-sucking insects, especially phloem-feeders, which represent a broad group of damaging pests, namely aphids, whiteflies, mealybugs, psyllids and leafhoppers. The last two groups include the majority of phytoplasma vectors, against which chemical insecticides are the main management strategies to counteract phytoplasma diseases.
This study successfully demonstrated the use of a plant-mediated delivery of dsRNAs to two sap-sucking leafhopper species, S. titanus and E. variegatus. For these species, microinjection of dsRNAs has been documented to produce an effective RNAi response (Abbà et al. 2019; Ripamonti et al. 2022a), but can be essentially only used for studies in laboratory conditions. Here, we showed that the soaking method, successful in dsRNA delivery to Diaphorina citri and Aedes aegypti (Killiny et al. 2014; Yu et al. 2017; Arshad et al. 2021), was ineffective in silencing target genes under our conditions. This was in line with data reported for Drosophila melanogaster larvae treated by soaking (Powell et al. 2017). We have no indication on the reason for the lack of silencing (degradation of dsRNAs or failure of delivery into treated insects), but, being ineffective, the soaking method was not further explored.
Petiole absorption has been a successful method in delivering dsRNAs systemically into plant xylem and apoplast (Ghosh et al. 2017; Dalakouras et al. 2018). It is noteworthy that, in general, phloem-feeders do not feed strictly on phloem only, but also probe xylem according to electro-penetrography data. In particular, this is true for the insect species under study, S. titanus (Chuche et al. 2017; Ripamonti et al. 2022b) and E. variegatus (M. Rossi preliminary unpublished data during feeding onto different plant hosts). This feature prompted us to attempt plant absorption as a delivery method of dsRNAs. Optimization of plant absorption conditions started with monitoring the movement of a coloured solution into plants and then into insects fed on treated plants. Microscopic observations showed that coloured solution reached the xylem elements of treated plants (broad bean, grapevine, and oat), in line with previous works on grapevine, apple and green bean (Ghosh et al. 2017; Dalakouras et al. 2018). Moreover, effective delivery to insects was confirmed by observation of stained guts and honeydews from insects fed on treated plants. Despite the petiole absorption was effective in the delivery of dsGFP in insects fed on treated whole plants, the use of detached leaves immersed in dsRNA solution increased the delivery efficiency to exposed insects from 40 to about 80%. This was the final chosen condition, since it allowed concentration of dsRNAs on a limited portion of plant tissue and reduction of the dsRNA quantity necessary to induce gene silencing.
Natterin and legumain of E. variegatus were chosen as target genes to be silenced for the initial optimization, due to their specific expression in insect gut tissues (Hartmann et al. 2018; Molina-Cruz et al. 2020), with the aim of obtaining a rapid delivery of dsRNAs to their target organ soon after their acquisition by feeding (Kunte et al. 2020). Analyses of these two transcripts on different dissected organs confirmed the strong tissue specific expression of both genes in E. variegatus guts. Furthermore, the developed dsRNAs targeting natterin and legumain produced effective gene silencing, following injection, confirming that their length and sequence base composition did not hamper their activity (Terenius et al. 2011; Miller et al. 2012; Joga et al. 2016). Interestingly, upon their acquisition by feeding on treated detached leaves, a significant reduction of specific transcripts was observed in insects from the second sampling date onwards, indicating that under our experimental conditions a time lag is necessary before the RNAi mechanism may be triggered, in line with the high variability of the temporal delay between treatment and the onset of RNAi-induced phenotype reported for different insect pests (Mehlhorn et al. 2020; Yoon et al. 2020; Silver et al. 2021).
Optimized conditions for detached leaf absorption were effective for both E. variegatus and S. titanus leafhopper species for the specific silencing of ATP synthase β gene, selected as systemically expressed and known to be involved in phytoplasma transmission (Galetto et al. 2011, 2021). Gene silencing by dsRNA uptake from detached leaves occurred with similar frequency as the staining incidence observed in dissected insect guts, upon feeding on leaves treated with colour solution. A significant reduction of specific transcript was observed for the two species, both for gut-specific genes, more likely to occur when dsRNAs are acquired through feeding, and for the systemically expressed ATP synthase β. In any case, a systemic distribution of acquired dsRNAs is demonstrated, as observed for many different pests (Tomoyasu et al. 2008; Joga et al. 2016; Vélez and Fishilevich 2018) and also for E. variegatus, in which dsATP are able to silence the specific transcript in tissues distal from the injection site (Galetto et al. 2021).
No effect on survival rate was observed in both species after feeding on dsRNAs targeting the three genes, at least for the time frame of our experiment. Proteins belonging to respiratory complexes such as ATP synthase usually show a slow turnover due to the formation of the mitochondrial respiratory complexes, which require over-transcription of the protein subunits to provide an adequate molecule supply for the assembly process (Bogenhagen and Haley 2020). Given that clustering of transmembrane cargo molecules has been shown to modulate the turnover kinetics (Morozova and Weiss 2010), a long turnover may also be hypothesized for legumain, which displays the presence of a predicted transmembrane domain (not shown).
In conclusion, a plant-mediated method to effectively deliver dsRNAs to phloem feeders has been proposed. Compared to microinjection, even if slightly less efficient (70 vs. 100%), dsRNA delivery by feeding is less time-consuming and allows processing larger set of specimens. Moreover, it is suitable for silencing every nymphal stage and does not require any specific device nor expertise. A short-term outcome of the proposed delivery method will be its exploitation in functional genomic studies, especially in the case of vector species of obligate fastidious pathogens, such as phytoplasmas or liberibacters. Indeed, functional studies to unveil mechanisms of interaction among pathogens and their hosts are hampered by difficulties in establishing axenic cultures. In this respect, RNAi is a useful tool to decipher the role of insect genes involved in transmission of fastidious pathogens (Sarkar and Ghanim 2020; Tang et al. 2020; Galetto et al. 2021; Arricau-Bouvery et al. 2023). This procedure is still not applicable to crop protection, but this work provides an important proof of concept: dsRNAs absorbed by plants can silence target genes of phloem-feeders fed on those plants. Although not straightforward, trunk injection of dsRNAs in woody plants may therefore become a feasible solution under field conditions to limit phloem-feeders. Nowadays, in many cases, synthetic insecticides are the unique current management strategy, although the chemical arsenal is gradually depleting in accordance with UE policies. This study addresses dsRNA delivery strategies to sap-sucking insects, a crucial issue for the effective exploitation of RNAi-based insecticides.
Author Contributions
MR, CM, DB, and LG designed the experiments, MR, SO, AF and LB performed the experiments, SA provided bioinformatic data, MR and LG performed the statistical analyses and drafted the manuscript, LG, DB and CM funded the work. All authors revised the manuscript draft and contributed to and accepted its final version.
References
Abbà S, Galetto L, Ripamonti M et al (2019) RNA interference of muscle actin and ATP synthase beta increases mortality of the phytoplasma vector Euscelidius variegatus. Pest Manag Sci 75:1425–1434. https://doi.org/10.1002/ps.5263
Abbà S, Rossi M, Vallino M et al (2022) Metatranscriptomic assessment of the microbial community associated with the Flavescence dorée phytoplasma insect vector Scaphoideus titanus. Front Microbiol 13:866523. https://doi.org/10.3389/fmicb.2022.866523
Arricau-Bouvery N, Duret S, Dubrana M-P et al (2018) Variable membrane protein A of flavescence dorée phytoplasma binds the midgut perimicrovillar membrane of Euscelidius variegatus and promotes adhesion to its epithelial cells. Appl Environ Microbiol 84:8–17. https://doi.org/10.1128/AEM.02487-17
Arricau-Bouvery N, Duret S, Dubrana M-P et al (2021) Interactions between the Flavescence dorée phytoplasma and its insect vector indicate lectin-type adhesion mediated by the adhesin VmpA. Sci Rep 11:11222. https://doi.org/10.1038/s41598-021-90809-z
Arricau-Bouvery N, Dubrana M-P, Canuto F et al (2023) Flavescence dorée phytoplasma enters insect cells by a clathrin-mediated endocytosis allowing infection of its insect vector. Sci Rep 13:2211. https://doi.org/10.1038/s41598-023-29341-1
Arshad F, Sharma A, Lu C, Gulia-Nuss M (2021) RNAi by soaking Aedes aegypti pupae in dsRNA. InSects 12:634. https://doi.org/10.3390/insects12070634
Baulcombe D (2004) RNA silencing in plants. Nature 431:356–363. https://doi.org/10.1038/nature02874
Baum JA, Bogaert T, Clinton W et al (2007) Control of coleopteran insect pests through RNA interference. Nat Biotechnol 25:1322–1326. https://doi.org/10.1038/nbt1359
Bogenhagen DF, Haley JD (2020) Pulse-chase SILAC–based analyses reveal selective oversynthesis and rapid turnover of mitochondrial protein components of respiratory complexes. J Biol Chem 295:2544–2554. https://doi.org/10.1074/jbc.RA119.011791
Bonning BC, Saleh M-C (2021) The interplay between viruses and RNAi pathways in insects. Annu Rev Entomol 66:61–79. https://doi.org/10.1146/annurev-ento-033020-090410
Carpane P, Wayadande A, Backus E et al (2011) Characterization and correlation of new electrical penetration graph waveforms for the corn leafhopper (Hemiptera: Cicadellidae). Ann Entomol Soc Am 104:515–525. https://doi.org/10.1603/AN10052
Caudwell A, Kuszala C, Larrue J, Bachelier J (1972) Transmission de la Flavescence dorée de la fève à la fève par des cicadelles des genres Euscelis et Euscelidius. Ann Phytopathol 1572:181–189
Christiaens O, Smagghe G (2014) The challenge of RNAi-mediated control of hemipterans. Curr Opin Insect Sci 6:15–21. https://doi.org/10.1016/j.cois.2014.09.012
Christiaens O, Petek M, Smagghe G, Taning CNT (2020a) The use of nanocarriers to improve the efficiency of RNAi-based pesticides in agriculture. In: Fraceto LF, de Vera Lucia SS, Castro RG, Ávila D, Oliveira HC, Lima R (eds) Nanopesticides: From Research and Development to Mechanisms of Action and Sustainable Use in Agriculture. Springer International Publishing, Cham, pp 49–68. https://doi.org/10.1007/978-3-030-44873-8_3
Christiaens O, Whyard S, Vélez AM, Smagghe G (2020b) Double-stranded RNA technology to control insect pests: current status and challenges. Front Plant Sci 11:451. https://doi.org/10.3389/fpls.2020.00451
Christiaens O, Sweet J, Dzhambazova T et al (2022) Implementation of RNAi-based arthropod pest control: environmental risks, potential for resistance and regulatory considerations. J Pest Sci 95:1–15. https://doi.org/10.1007/s10340-021-01439-3
Chuche J, Thiéry D (2014) Biology and ecology of the Flavescence dorée vector Scaphoideus titanus: a review. Agron Sustain Dev 34:381–403. https://doi.org/10.1007/s13593-014-0208-7
Chuche J, Sauvion N, Thiéry D (2017) Mixed xylem and phloem sap ingestion in sheath-feeders as normal dietary behavior: evidence from the leafhopper Scaphoideus titanus. J Insect Physiol 102:62–72. https://doi.org/10.1016/j.jinsphys.2017.01.014
Coleman AD, Wouters RHM, Mugford ST, Hogenhout SA (2015) Persistence and transgenerational effect of plant-mediated RNAi in aphids. J Exp Bot 66:541–548. https://doi.org/10.1093/jxb/eru450
Dalakouras A, Jarausch W, Buchholz G et al (2018) Delivery of hairpin RNAs and small RNAs into woody and herbaceous plants by trunk injection and petiole absorption. Front Plant Sci 9:1253. https://doi.org/10.3389/fpls.2018.01253
Debonneville C, Mandelli L, Brodard J et al (2022) The complete genome of the “Flavescence dorée” phytoplasma reveals characteristics of low genome plasticity. Biology 11:953. https://doi.org/10.3390/biology11070953
Dietz-Pfeilstetter A, Mendelsohn M, Gathmann A, Klinkenbuß D (2021) Considerations and regulatory approaches in the USA and in the EU for dsRNA-based externally applied pesticides for plant protection. Front Plant Sci 12:682387. https://doi.org/10.3389/fpls.2021.682387
EFSA Panel on Plant Health PLH (2014) Scientific opinion on pest categorisation of grapevine Flavescence dorée. EFSA J 12: 3851 https://doi.org/10.2903/j.efsa.2014.3851
EFSA (European Food Safety Authority), Baker R, Gilioli G, et al (2019) Grapevine Flavescence dorée pest report and datasheet to support ranking of EU candidate priority pests. https://zenodo.org/record/2789595
Fire A, Xu S, Montgomery MK et al (1998) Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 391:806–811. https://doi.org/10.1038/35888
Galetto L, Bosco D, Balestrini R et al (2011) The major antigenic membrane protein of “Candidatus Phytoplasma asteris” selectively interacts with ATP synthase and actin of leafhopper vectors. PLoS ONE 6:e22571. https://doi.org/10.1371/journal.pone.0022571
Galetto L, Bosco D, Marzachì C (2013) Selection of reference genes from two leafhopper species challenged by phytoplasma infection, for gene expression studies by RT-qPCR. BMC Res Notes 6:409. https://doi.org/10.1186/1756-0500-6-409
Galetto L, Abbà S, Rossi M et al (2018) Two phytoplasmas elicit different responses in the insect vector Euscelidius variegatus Kirschbaum. Infect Immun 86(86):10–128. https://doi.org/10.1128/IAI.00042-18
Galetto L, Abbà S, Rossi M et al (2021) Silencing of ATP synthase β reduces phytoplasma multiplication in a leafhopper vector. J Insect Physiol 128:104176. https://doi.org/10.1016/j.jinsphys.2020.104176
Ghosh SKB, Hunter WB, Park AL, Gundersen-Rindal DE (2017) Double strand RNA delivery system for plant-sap-feeding insects. PLoS ONE 12:e0171861. https://doi.org/10.1371/journal.pone.0171861
Hartmann D, Šíma R, Konvičková J et al (2018) Multiple legumain isoenzymes in ticks. Int J Parasitol 48:167–178. https://doi.org/10.1016/j.ijpara.2017.08.011
Huvenne H, Smagghe G (2010) Mechanisms of dsRNA uptake in insects and potential of RNAi for pest control: a review. J Insect Physiol 56:227–235. https://doi.org/10.1016/j.jinsphys.2009.10.004
Jain RG, Fletcher SJ, Manzie N et al (2022) Foliar application of clay-delivered RNA interference for whitefly control. Nat Plants 8:535–548. https://doi.org/10.1038/s41477-022-01152-8
Joga MR, Zotti MJ, Smagghe G, Christiaens O (2016) RNAi efficiency, systemic properties, and novel delivery methods for pest insect control: what we know so far. Front Physiol 7:553. https://doi.org/10.3389/fphys.2016.00553
Killiny N, Hajeri S, Tiwari S et al (2014) Double-stranded RNA uptake through topical application mediates silencing of five CYP4 genes and suppresses insecticide resistance in Diaphorina citri. PLoS ONE 9:e110536. https://doi.org/10.1371/journal.pone.0110536
Kunte N, McGraw E, Bell S et al (2020) Prospects, challenges and current status of RNAi through insect feeding. Pest Manag Sci 76:26–41. https://doi.org/10.1002/ps.5588
Lett J-M, Granier M, Grondin M et al (2001) Electrical penetration graphs from Cicadulina mbila on maize, the fine structure of its stylet pathways and consequences for virus transmission efficiency. Entomol Exp Appl 101:93–109. https://doi.org/10.1046/j.1570-7458.2001.00895.x
Li H, Bowling AJ, Gandra P et al (2018) Systemic RNAi in Western corn rootworm, Diabrotica virgifera virgifera, does not involve transitive pathways: no transitive RNAi in Western corn rootworm. Insect Sci 25:45–56. https://doi.org/10.1111/1744-7917.12382
Lindbo JA (2007) TRBO: a high-efficiency Tobacco Mosaic Virus RNA-based overexpression vector. Plant Physiol 145:1232–1240. https://doi.org/10.1104/pp.107.106377
Liu S, Jaouannet M, Dempsey DA et al (2020) RNA-based technologies for insect control in plant production. Biotechnol Adv 39:107463. https://doi.org/10.1016/j.biotechadv.2019.107463
Malembic-Maher S, Desqué D, Khalil D et al (2020) When a Palearctic bacterium meets a Nearctic insect vector: genetic and ecological insights into the emergence of the grapevine Flavescence dorée epidemics in Europe. PLOS Pathog 16:e1007967. https://doi.org/10.1371/journal.ppat.1007967
Mehlhorn SG, Geibel S, Bucher G, Nauen R (2020) Profiling of RNAi sensitivity after foliar dsRNA exposure in different European populations of Colorado potato beetle reveals a robust response with minor variability. Pestic Biochem Physiol 166:104569. https://doi.org/10.1016/j.pestbp.2020.104569
Mezzetti B, Smagghe G, Arpaia S et al (2020) RNAi: What is its position in agriculture? J Pest Sci 93:1125–1130. https://doi.org/10.1007/s10340-020-01238-2
Miller SC, Brown SJ, Tomoyasu Y (2008) Larval RNAi in Drosophila? Dev Genes Evol 218:505–510. https://doi.org/10.1007/s00427-008-0238-8
Miller SC, Miyata K, Brown SJ, Tomoyasu Y (2012) Dissecting systemic RNA interference in the red flour beetle Tribolium castaneum: parameters affecting the efficiency of RNAi. PLoS ONE 7:e47431. https://doi.org/10.1371/journal.pone.0047431
Molina-Cruz A, Canepa GE, Alves e Silva TL et al (2020) Plasmodium falciparum evades immunity of anopheline mosquitoes by interacting with a Pfs47 midgut receptor. Proc Natl Acad Sci 117:2597–2605. https://doi.org/10.1073/pnas.1917042117
Morozova D, Weiss M (2010) On the role of acylation of transmembrane proteins. Biophys J 98:800–804. https://doi.org/10.1016/j.bpj.2009.11.014
OECD (2020) Considerations for the environmental risk assessment of the application of sprayed or externally applied dsRNA-based pesticides. Ser Pestic 104:ENV/JM/MONO(2020)26
Oliveira MJRA, Roriz M, Vasconcelos MW et al (2019) Conventional and novel approaches for managing “Flavescence dorée” in grapevine: knowledge gaps and future prospects. Plant Pathol 68:3–17. https://doi.org/10.1111/ppa.12938
Oshima K, Maejima K, Namba S (2013) Genomic and evolutionary aspects of phytoplasmas. Front Microbiol 4:230. https://doi.org/10.3389/fmicb.2013.00230
Ottati S, Chiapello M, Galetto L et al (2020) New viral sequences identified in the Flavescence dorée phytoplasma vector Scaphoideus titanus. Viruses 12:287. https://doi.org/10.3390/v12030287
Palli SR (2014) RNA interference in Colorado potato beetle: steps toward development of dsRNA as a commercial insecticide. Curr Opin Insect Sci 6:1–8. https://doi.org/10.1016/j.cois.2014.09.011
Papadopoulou N, Devos Y, Álvarez-Alfageme F et al (2020) Risk assessment considerations for genetically modified RNAi plants: EFSA’s activities and perspective. Front Plant Sci 11:445. https://doi.org/10.3389/fpls.2020.00445
Pinzón N, Bertrand S, Subirana L et al (2019) Functional lability of RNA-dependent RNA polymerases in animals. PLOS Genet 15:e1007915. https://doi.org/10.1371/journal.pgen.1007915
Powell M, Pyati P, Cao M et al (2017) Insecticidal effects of dsRNA targeting the Diap1 gene in dipteran pests. Sci Rep 7:15147. https://doi.org/10.1038/s41598-017-15534-y
Pugsley CE, Isaac RE, Warren NJ, Cayre OJ (2021) Recent advances in engineered nanoparticles for RNAi-mediated crop protection against insect pests. Front Agron 3:652981. https://doi.org/10.3389/fagro.2021.652981
Ripamonti M, Cerone L, Abbà S et al (2022a) Silencing of ATP synthase β impairs egg development in the leafhopper Scaphoideus titanus, vector of the phytoplasma associated with grapevine Flavescence dorée. Int J Mol Sci 23:765. https://doi.org/10.3390/ijms23020765
Ripamonti M, Maron F, Cornara D et al (2022b) Leafhopper feeding behaviour on three grapevine cultivars with different susceptibilities to Flavescence dorée. J Insect Physiol 137:104366. https://doi.org/10.1016/j.jinsphys.2022.104366
Santos D, Mingels L, Vogel E et al (2019) Generation of virus- and dsRNA-derived siRNAs with species-dependent length in insects. Viruses 11:738. https://doi.org/10.3390/v11080738
Sarkar P, Ghanim M (2020) Unravelling the pathogenesis and molecular interactions of Liberibacter phytopathogens with their psyllid vectors. Agronomy 10:1132. https://doi.org/10.3390/agronomy10081132
Silver K, Cooper AM, Zhu KY (2021) Strategies for enhancing the efficiency of RNA interference in insects. Pest Manag Sci 77:2645–2658. https://doi.org/10.1002/ps.6277
Stafford CA, Walker GP (2009) Characterization and correlation of DC electrical penetration graph waveforms with feeding behavior of beet leafhopper, Circulifer tenellus. Entomol Exp Appl 130:113–129. https://doi.org/10.1111/j.1570-7458.2008.00812.x
Tang X-T, Fortuna K, Mendoza Herrera A, Tamborindeguy C (2020) Liberibacter, a preemptive bacterium: apoptotic response repression in the host gut at the early infection to facilitate its acquisition and transmission. Front Microbiol 11:589509. https://doi.org/10.3389/fmicb.2020.589509
Terenius O, Papanicolaou A, Garbutt JS et al (2011) RNA interference in Lepidoptera: an overview of successful and unsuccessful studies and implications for experimental design. J Insect Physiol 57:231–245. https://doi.org/10.1016/j.jinsphys.2010.11.006
Tomoyasu Y, Miller SC, Tomita S et al (2008) Exploring systemic RNA interference in insects: a genome-wide survey for RNAi genes in Tribolium. Genome Biol 9:R10. https://doi.org/10.1186/gb-2008-9-1-r10
Tonkyn DW, Whitcomb RF (1987) Feeding strategies and the guild concept among vascular feeding insects and microorganisms. In: Lal R, Stewart BA (eds) Soil Restoration. Springer, New York, pp 179–199
Tramontini S, Delbianco A, Vos S (2020) Pest survey card on Flavescence dorée phytoplasma and its vector Scaphoideus titanus. EFSA Support Publ. https://doi.org/10.2903/sp.efsa.2020.EN-1909
Trębicki P, Tjallingii WF, Harding RM et al (2012) EPG monitoring of the probing behaviour of the common brown leafhopper Orosius orientalis on artificial diet and selected host plants. Arthropod-Plant Interact 6:405–415. https://doi.org/10.1007/s11829-012-9192-5
Trivellone RM, Angelini E et al (2019) Evidence suggesting interactions between immunodominant membrane protein Imp of Flavescence dorée phytoplasma and protein extracts from distantly related insect species. J Appl Microbiol 127:1801–1813. https://doi.org/10.1111/jam.14445
Vélez AM, Fishilevich E (2018) The mysteries of insect RNAi: a focus on dsRNA uptake and transport. Pestic Biochem Physiol 151:25–31. https://doi.org/10.1016/j.pestbp.2018.08.005
Wayadande A (1994) Electronic monitoring of leafhoppers and planthoppers: feeding behavior and application in host-plant resistance studies. In: Ellsbury MM, Backus EA, Ullman DL (eds) History, development, and application of AC electronic insect feeding monitors, Thomas Say Publications in Entomology, Entomological Society of America. Maryland, USA, pp 96–105
Wei D, Wang S, Niu J (2023) RNAi-based pesticides: current knowledge and potential applications for Integrated Pest Management. Entomol Gen 43:1–4. https://doi.org/10.1127/entomologia/2023/1988
Whyard S, Singh AD, Wong S (2009) Ingested double-stranded RNAs can act as species-specific insecticides. Insect Biochem Mol Biol 39:824–832. https://doi.org/10.1016/j.ibmb.2009.09.007
Yoon J-S, Tian H, McMullen JG et al (2020) Candidate genetic determinants of intraspecific variation in pea aphid susceptibility to RNA interference. Insect Biochem Mol Biol 123:103408. https://doi.org/10.1016/j.ibmb.2020.103408
Yu N, Christiaens O, Liu J et al (2013) Delivery of dsRNA for RNAi in insects: an overview and future directions. Insect Sci 20:4–14. https://doi.org/10.1111/j.1744-7917.2012.01534.x
Yu X-D, Liu Z-C, Huang S-L et al (2016) RNAi-mediated plant protection against aphids. Pest Manag Sci 72:1090–1098. https://doi.org/10.1002/ps.4258
Yu X, Gowda S, Killiny N (2017) Double-stranded RNA delivery through soaking mediates silencing of the muscle protein and increases mortality to the Asian citrus psyllid, Diaphorina citri. Pest Manag Sci 73:1846–1853. https://doi.org/10.1002/ps.4549
Zhu KY, Palli SR (2020) Mechanisms, applications, and challenges of insect RNA interference. Annu Rev Entomol 65:293–311. https://doi.org/10.1146/annurev-ento-011019-025224
Zhu F, Xu J, Palli R et al (2011) Ingested RNA interference for managing the populations of the Colorado potato beetle, Leptinotarsa decemlineata. Pest Manag Sci 67:175–182. https://doi.org/10.1002/ps.2048
Zong J, Yao X, Yin J et al (2009) Evolution of the RNA-dependent RNA polymerase (RdRP) genes: duplications and possible losses before and after the divergence of major eukaryotic groups. Gene 447:29–39. https://doi.org/10.1016/j.gene.2009.07.004
Acknowledgements
The authors wish to thank Flavio Veratti for his skilled technical help, Elena Zocca for her indispensable help in plant production and greenhouse assistance, Sabrina Palmano and Marta Vallino for their constructive advices and comments to the manuscript.
Funding
Open access funding provided by Consiglio Nazionale Delle Ricerche (CNR) within the CRUI-CARE Agreement. This work was part of the FOotSTEP and ReSet Projects, funded by Fondazione Cassa di Risparmio di Torino, Turin (Italy) (Grant Numbers 2018–0678 and 2022.1790).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors have no relevant financial or non-financial interests to disclose.
Additional information
Communicated by Subba Palli.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Rossi, M., Ottati, S., Bucci, L. et al. Lab-scale method for plant-mediated delivery of dsRNAs to phloem-feeding leafhoppers. J Pest Sci 97, 455–467 (2024). https://doi.org/10.1007/s10340-023-01670-0
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10340-023-01670-0