Abstract
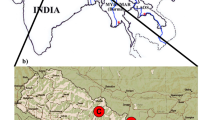
The Tonkin snub-nosed monkey (Rhinopithecus avunculus) with a global population of <250 is listed as critically endangered. It is endemic to northeastern Vietnam and was feared extinct until its rediscovery in 1989. The largest single population of R. avunculus consists of 125–130 individuals in an area of forest called Khau Ca in Ha Giang Province. We used non-invasively collected fecal samples to establish the amount of genetic diversity in this population based on mitochondrial information. We amplified and sequenced a 467- to 650-bp section of the hypervariable region I (HVI) of the mitochondrial D-loop for 201 samples and reconstructed the full mitochondrial genomes for five samples based on metagenomic data. All 201 HVI sequences were identical and no variability was found in the five mitochondrial genomes. Our results highlight the immediate need for a comprehensive assessment of the genetic diversity of all populations of R. avunculus based on mitochondrial and nuclear markers. The latter need to be developed for this species.


Similar content being viewed by others
References
Ang A, Srivathsan A, Md-Zain BM, Ismail MRB, Meier R (2012) Low genetic variability in the recovering urban banded leaf monkey population of Singapore. Raffles B Zool 60:589–594
Baillie JEM, Butcher ER (2012) Priceless or worthless? The world’s most threatened species. Zoological Society of London, UK
Bogler AM, Lohse M, Usadel B (2014) Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30(15):2114–2120
Castro-Prieto A, Wachter B, Sommer S (2011) Cheetah paradigm revisited: MHC diversity in the world’s largest free-ranging population. Mol Biol Evol 28(4):1455–1468
Faircloth BC, Glenn TC (2014) Protocol: Preparation of an AMPure XP substitute (AKA Serapure). doi:10.6079/J9MW2F26
Fooden J (1996) Zoogeography of Vietnamese primates. Int J Primatol 17(5):845–899
Garrison E, Marth G (2012) Haplotype-based variant detection from short-read sequencing. arXiv:1207.3907 [q-bio.GN]
Katoh M, Kuma M (2002) MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res 30:3059–3066
Kolleck J, Yang M, Zinner D, Roos C (2013) Genetic diversity in endangered Guizhou snub-nosed monkeys (Rhinopithecus brelichi): contrasting results from microsatellite and mitochondrial DNA data. PLoS One 8(8):e73647. doi:10.1371/journal.pone.0073647
Kutty SN, Bernasconi MV, Šifner F, Meier R (2007) Sensitivity analysis, molecular systematics and natural history evolution of Scathophagidae (Diptera: Cyclorrhapha: Calyptratae). Cladistics 23:64–83
Lande R, Barrowclough G (1987) Effective population size, genetic variation, and their use in population management. In: Soulé ME (ed) Viable populations for conservation. Cambridge University Press, New York, pp 87–123
Langmead B, Salzberg SL (2012) Fast gapped-read alignment with Bowtie 2. Nat Methods 9:357–359
Le KQ (2002) Distribution and conservation of Tonkin snub-nosed monkey (Rhinopithecus avunculus) in Du Gia Nature Reserve, Ha Giang Province, Northeast Vietnam. Fauna and Flora International-Indochina Programme
Le KQ, Dong TH, Nadler T (2015) Tonkin snub-nosed monkey Rhinopithecus avunculus (Dollman 1912). In: Schwitzer C, Mittermeier RA, Rylands AB, Chiozza F, Williamson EA, Wallis J, Cotton A (eds) Primates in peril: the world’s 25 most endangered primates 2014–2016. IUCN SSC PSG, IPS, CI, and Bristol Zoological Society, Arlington, pp 58–59
Leese F, Held C (2011) Analysing intraspecific genetic variation: a practical guide using mitochondrial DNA and microsatellites. In: Held C, Koenemann S, Schubart C (eds) Phylogeography and population genetics in Crustacea. CRC, Boca Raton, pp 3–30
Li H, Durbin R (2009) Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25(14):1754–1760
Lopez JV, Culver M, Stephens JC, Johnson WE, O’Brien SJ (1997) Rates of nuclear and cytoplasmic mitochondrial DNA sequence divergence in mammals. Mol Biol Evol 14:277–286
Ma C, Huang ZP, Zhao XF, Zhang LX, Sun WM, Scott MB, Wang XW et al (2014) Distribution and conservation status of Rhinopithecus strykeri in China. Primates 55:377–382
Meier R, Wong W, Srivathsan A, Foo M (2016) $1 DNA barcodes for reconstructing complex phenomes and finding rare species in specimen-rich samples. Cladistics 32:100–110
Miller W, Hayes VM, Ratan A, Petersen DC, Wittekindt NE, Miller J, Walenz B et al (2011) Genetic diversity and population structure of the endangered marsupial Sarcophilus harrisii (Tasmanian devil). PNAS 108(30):12348–12353
Mittermeier RA, Cheney DL (1987) Conservation of primates and their habitats. In: Smuts BB, Cheney DL, Seyfarth RM, Wrangham RW, Strushsaker TT (eds) Primate societies. University of Chicago Press, Chicago, pp 477–490
Nadler T (2014) Habitat disturbance and loss, and the primates of Vietnam. In: Nadler T, Brockman D (eds) Primates of Vietnam. Endangered Primate Rescue Center, Cuc Phuong National Park, Vietnam, pp 55–60
Nsubuga AM, Robbins MM, Roeder AD, Morin PA, Boesch C, Vigilant L (2004) Factors affecting amount of genomic DNA extracted from ape faeces and the identification of an improved sample storage method. Mol Ecol 13:2089–2094
Osterholz M, Walter L, Roos C (2008) Phylogenetic position of the langur genera Semnopithecus and Trachypithecus among Asian colobines, and genus affiliations of their species groups. BMC Evol Biol 8:58–69
Ratajszczak R, Can Dang Ngoc, Nhat Pham (1992) A survey for Tonkin snub-nosed monkey (Rhinopithecus avunculus) in the North Vietnam. FFI Preservation Society and WWF International, London and Gland
Rohland N, Reich D (2012) Cost-effective, high-throughput DNA sequencing libraries for multiplexed target capture. Genome Res 22:939–946
Roos C, Thanh Vu Ngoc, Walter L, Nadler T (2007) Molecular systematics of Indochinese primates. Vietnam J Primatol 1:41–53
Roos C, Zinner D, Kubatko LS, Schwarz C, Yang M, Meyer D, Nash SD, Xing J, Batzer MA, Brameier M, Leendertz FH, Ziegler T, Perwitasari-Farajallah D, Nadler T, Walter L, Osterholz M (2011) Nuclear versus mitochondrial DNA: evidence for hybridization in colobine monkeys. BMC Evol Biol 11:77
Spielman D, Brook BW, Briscoe DA, Frankham R (2004) Does inbreeding and loss of genetic diversity decrease disease resistance? Conserv Genet 5:439–448
Srivathsan A, Sha J, Vogler AP, Meier R (2015) Comparing the effectiveness of metagenomics and metabarcoding for diet analysis of a leaf-feeding monkey (Pygathrix nemaeus). Mol Ecol Res 15(2):250–261
Srivathsan A, Ang A, Vogler AP, Meier R (2016) Fecal metagenomics for the simultaneous assessment of diet, parasites, and population genetics of an understudied primate. Front Zool 13:17. doi:10.1186/s12983-016-0150-4
Wisely SM, Buskirk SW, Fleming MA, McDonald DB, Ostrander EA (2002) Genetic diversity and fitness in black-footed ferrets before and during a bottleneck. J Hered 93:231–237
Yang M, Yang Y, Cui D, Fickenscher G, Zinner D, Roos C, Brameier M (2012) Population genetic structure of Guizhou snub-nosed monkeys (Rhinopithecus brelichi) as inferred from mitochondrial control region sequences, and comparison with R. roxellana and R. bieti. Am J Phys Anthropol 147:1–10
Yu L, Wang X, Ting N, Zhang Y (2011) Mitogenomic analysis of Chinese snub-nosed monkeys: evidence of positive selection in NADH dehydrogenase genes in high-altitude adaptation. Mitochondrion 11(3):497–503
Zhou X, Wang B, Pan Q, Zhang J, Kumar S, Sun X, Liu Z et al (2014) Whole-genome sequencing of the snub-nosed monkey provides insights into folivory and evolutionary history. Nat Genet 46:1303–1310
Acknowledgments
We thank everyone for their help, particularly the management board of the Tonkin Snub-nosed Monkey Species Habitat Conservation Area at Khau Ca; Mr Hoang Van Tue, Vietnam Administration of Forestry, Southern Institute of Ecology; Mr Dan Van Khoan, his family and his team; and the Evolutionary Biology Lab (National University of Singapore). We would also like to thank AIT Biotech for facilitating the NGS sequencing.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
We followed the Code of Best Practices for Field Primatology (2014) and the ethical guidelines of the University of Colorado Boulder and the National University of Singapore. Research permits were obtained and we followed the rules and regulations of the Forest Protection Department of Ha Giang, the Protected Area Management Board of Khau Ca, and relevant Vietnamese authorities (permit no. 2039/TCLN-BTTN).
Funding
This research was funded by Wildlife Reserves Singapore, the Mohamed bin Zayed Species Conservation Fund, the Beverly Sears Graduate Student Research Grant, the Scott Ferris Graduate Research Award, the Department of Anthropology Pre-Dissertation Grant, SEABIG (NUS) R-154-000-648-646 and R-154-000-648-733, and Dr Vilma D’Rozario.
Conflict of interest
The authors declare that they have no conflict of interest and gave final approval for publication.
Electronic supplementary material
Below is the link to the electronic supplementary material.
About this article
Cite this article
Ang, A., Srivathsan, A., Meier, R. et al. No evidence for mitochondrial genetic variability in the largest population of critically endangered Tonkin snub-nosed monkeys in Vietnam. Primates 57, 449–453 (2016). https://doi.org/10.1007/s10329-016-0571-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10329-016-0571-x