Abstract
This paper aims to solve the arterial input function (AIF) determination in dynamic contrast-enhanced MRI (DCE-MRI), an important linear ill-posed inverse problem, using the maximum entropy technique (MET) and regularization functionals. In addition, estimating the pharmacokinetic parameters from a DCE-MR image investigations is an urgent need to obtain the precise information about the AIF–the concentration of the contrast agent on the left ventricular blood pool measured over time. For this reason, the main idea is to show how to find a unique solution of linear system of equations generally in the form of \(y=Ax+b,\) named an ill-conditioned linear system of equations after discretization of the integral equations, which appear in different tomographic image restoration and reconstruction issues. Here, a new algorithm is described to estimate an appropriate probability distribution function for AIF according to the MET and regularization functionals for the contrast agent concentration when applying Bayesian estimation approach to estimate two different pharmacokinetic parameters. Moreover, by using the proposed approach when analyzing simulated and real datasets of the breast tumors according to pharmacokinetic factors, it indicates that using Bayesian inference—that infer the uncertainties of the computed solutions, and specific knowledge of the noise and errors—combined with the regularization functional of the maximum entropy problem, improved the convergence behavior and led to more consistent morphological and functional statistics and results. Finally, in comparison to the proposed exponential distribution based on MET and Newton’s method, or Weibull distribution via the MET and teaching–learning-based optimization (MET/TLBO) in the previous studies, the family of Gamma and Erlang distributions estimated by the new algorithm are more appropriate and robust AIFs.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
As a fast and noninvasive approach, dynamic contrast-enhanced magnetic resonance imaging (DCE-MRI) is widely used to analyze to quantitatively analyze perfusion in soft tissues in various clinical applications. These include the detection, characterization, and monitoring of different diseases for therapeutic purposes [1,2,3,4,5,6]. Typically, pharmacokinetic models are used in the quantitative analysis of DCE-MR images. For the DCE-MRI scan, an extracellular contrast agent with a low molecular weight such as gadolinium diethylenetriaminepentaacetic acid, Gd-DTPA, is injected. The in vivo concentration of the contrast agent (CA) in tissue over time is measured using T1-weighted images. Several pharmacokinetic models have been developed for the characterization of the signal intensity change over time. These models allow to quantify local physiologic features of the tissue, known as pharmacokinetic parameters [7].
Among pharmacokinetic models, the two-compartment model is the most popular [7]. In this model, the change in the contrast agent concentration is attributed to its transfer between two compartments: the blood plasma and the extravascular extracellular space (EES) of the tissue. The pharmacokinetic model can be determined as solution to an ordinary-differential equation (ODE) describing the exchange between the compartments [8,9,10,11,12,13,14,15].
An important term in the pharmacokinetic model is the arterial input function (AIF), that is CA concentration in the left ventricular blood pool over time. Despite the fact that the AIF itself has no clinical relevance, its precise calculation is of particular interest for proper estimation of pharmacokinetic parameters [16, 17]. Given the strong dependence of the determined rate constants on the AIF [18,19,20,21,22], their quantification in an absolute and reliable manner requires a precise measurement.
However, in many cases the direct measurement of the AIF from DCE-MRI images is not possible, as no large vessel is in the field of view, for example in breast scans. As a replacement, it has been proposed to use a simplified method such as a population averaged AIF. These AIFs include bi-exponential functions with parameters obtained by [23, 24] or a mix of the two Gaussian with an exponential [8,9,10,11,12,13,14,15]. The literature is split over the efficiency of population averaged AIF, with some authors reporting its ability to adequately estimating pharmacokinetic parameters [25, 26], while others raise concerns [27, 28].
In recent years, a couple of models have been developed to estimate the AIF from DCE-MRI scans without larger vessels in the field of view. The aim is to estimate the AIF together with the corresponding pharmacokinetic parameters from the CA signal over time [29,30,31,32]. For partial and fully automated AIF estimation, several different techniques have been proposed. Fan et al. [33] attempted to extract the AIF with a cluster method, using a manually marked region of interest (ROI). Reishofer et al. [34] proposed AIF extraction using classification based on criteria involving inherent arterial input features including an early bolus arrival and fast passage, as well as a high-contrast agent concentration.
In this paper, we propose a novel method for estimating the probability density function (PDF) of the AIF directly from measured concentration-time curves in enhanced tissues. Statistically speaking, the determination of the AIF from DCE-MRI scans is seen as the determination of the PDF of sample data. From a conceptual perspective, the choice of the type of distribution is an open problem. When available information is limited, e.g., sample size is small and/or has lower–order moments, an approach based on the maximum entropy (ME) principle can be the solution. Using all available data the maximum entropy distribution is the estimation with the smallest bias. Nevertheless, applying ME has a number of theoretical and practical restrictions.
Many nonparametric and parametric techniques have been proposed to estimate the probability density function of a random variable from observations. The maximum entropy technique (MET) is a widely used method to estimate and determine the probability density, with known high accuracy and efficiency. In MET an optimization problem is solved to obtain the unknown density. Jaynes [35] proposed the ME principle as a statistical inference method and stated that by employing this principle, a probability density function is selected that corresponds to the available knowledge and provides no unwarranted information. In this regard, among probability density functions meeting some constraints, the one with smaller entropy has more information, hence less uncertainty [35,36,37,38]. Over the past decade, there has been an extensive application of entropy maximization or similar approaches, including the determination of macromolecular structures and interactions [39,40,41,42,43,44,45,46,47,48,49,50] and the inference of signaling [51,52,53] and regulatory networks [54, 55] as well as the coding organization in neural populations [56,57,58,59,60,61,62,63,64,65] according to the analysis of DNA sequences (e.g., the identification of specific binding sites) [65,66,67,68,69,70,71]. In addition, MET is an often used tool for image reconstruction. This includes applications in radio astronomical interferometry, dealing, on a daily basis, with images with large dynamic ranges and up to one million pixels [72,73,74,75].
In a previous work, we proposed using a combination of MET and Newton’s method for AIF estimation and maximum a posterior (MAP) for estimation of the pharmacokinetic parameters [76]. In another study, we proposed two enhanced algorithms to estimate the AIF as a combination of Bayesian inference and optimization techniques. The first algorithm combines MET, teaching–learning-based optimization (TLBO) to assess the performance of observer in the classification tasks with existing data, and Bayesian methods to estimate the pharmacokinetic parameters [77]. Similar to other algorithms inspired by nature, TLBO is also a population-based approach and employs a population of solutions to obtain a global result [31, 78]. The second algorithm is the combination of MET, a concave optimization method, and the general regularization approach. In the present work, we propose to regularize the ME problem and the ill-conditioned linear system of equations in the pharmacokinetic model. In general, regularization is an appropriate method to find stable solution to ill-posed inverse problems [79].
The first proposed algorithm uses the Weibull distribution as most robust model for the AIF. In the proposed improved algorithm, via regularization the family of Gamma distributions is the best solution of the ME problem . To estimate physiological parameters, extensive investigation was performed on empirical data, such that a better understanding of the performance of the proposed method could be obtained. The previously analyzed data were provided by Paul Strickland Scanner Center, Mount Vernon Hospital, Northwood, UK [7]. The data were acquired according to the recommendations of [80]. Informed consent was obtained from all patients.
The following sections are organized as follows. “Methodology” gives a brief description of the methodology. The pharmacokinetic model for the analysis of DCE-MRI data is described. Then the MET for the ill-posed inverse problem is developed, following by the regularization of the ME problem. The TLBO algorithm is presented, followed by the Bayesian approach. In addition, some characteristics and flowchart of the proposed algorithm are also provided. In “Numerical Experiment”, the developed method is used to analyze the in vivo DCE-MRI data. “Discussion and Conclusions” provides concluding remarks.
Methodology
We propose a novel algorithm combining the MET with regularization functionals, enabling us to estimate the PDF of the AIF along with the pharmacokinetic parameters in a similar method in a Bayesian framework. Compared to previous methods, the proposed algorithm does not consist of several phases, decreasing the computation time of the estimation. The proposed algorithm is a robust combination of MET, regularization, and the Bayesian estimation approach.
Basic Model
Pharmacokinetic Model
Here, the popular pharmacokinetic model [81] is considered. The assumption of this model is that the CA resides in two compartments of the tissue, the vascular space and the extracellular extravascular space (EES), with exchange of CA between these two compartments. The exchange of CA in the tissue of interest (\(C_{T}(t)\)) can be described via an ODE [8,9,10,11,12,13,14,15,16],
where \(C_{p} (t)\) gives the concentration of CA in the vascular blood pool, that is the AIF, \(K_{a}\) and \(K_{b}\) are constants quantifying the CA exchange rate between plasma and extravascular-extracellular space (EES), respectively. With initial condition \(C_{p} (0)=0\), the integration form of Eq. (1) is as follows,
Equation (2) is a commonly used in many applications [24]. Murase [82] suggested another solution of Eq. (1) using discretization:
In matrix form, Eq. (3) is
in which the matrix \(\vec{A}_{ \times 2} = \{A(1), \ldots , A(n)\}'\) with \(I=1,2,...,n:\) includes n rows which are defined as \({A(I)=(\int _{0}^{t_{I} } C_{p} (s)ds, -\int _{0}^{t_{I} } C_{T} (s)ds),}\) and \(\vec{K}=\left( {K_{a} }, {K_{b}}\right) '\) and \(\vec{C}_{T}=\left( {C_{T}(t_{1})},{C_{T} (t_{2} )}, {\ldots }, {C_{T} (t_{n})} \right) '.\) The following linear system of equations arising in various tomographic image restoration and reconstruction problems is considered:
where \(b _{i} \mathrm{\sim }N(0,\sigma ^{2})\) and \(Y_{tis}(t_{i})\) are, respectively, the measured concentration in tissue at time \(t_{i}\) and the measured uncertainty (noise), considered to be additive, white, centered, Gaussian and independent of K [83].
The ill-posed inverse problem (IPP) Eq. (5) can be simplified to estimate K subject to A and Y. When the forward solution is determined, the important step is to estimate \(\hat{K}\) such that K optimizes the related measures, like the least square criterion, \(J(k)= {\Vert Y-AK \Vert }^{2}\). However, the model might fit to the data, but due to the ill-posedness of the linear problems, it may not have desired properties [79]. To this end, one can consider some initial prior information regarding errors and the unknowns K. The problem can then be handled using general regularization theory and by application of the statistical inference. Two different strategies can be used for this, either information theory and entropy, or Bayesian inference [79].
Regularization Methods
Regularization is an appropriate method to find a unique and stable solution to the IIP in Eq. (5), [79]. There are two issues at hand. The first one is that \(Y = AK\) has more than one solution and there is a need to know more conditions, for example \(\bigtriangleup (K,q)\) to choose that unique solution by
where q is a prior solution and \(\bigtriangleup\) a distance measure. The Lagrangian approach [79, 84] has been described as the best method to solve this. With the Lagrangian \(L(K,q)=\bigtriangleup (K,q)+ \lambda ^{t}(Y-AK)\) on can estimate \((\hat{\lambda }, \hat{K})\) via
or another solution by
\(\bigtriangleup (Y,AK)\) is a measure of distance between Y and AK. In here, the \(\bigtriangleup (Y,AK)={\Vert Y-AK \Vert }^{2}\) is least square (LS) criterion. Obviously, if \(\hat{K}\) satisfies \(A^{t}A\hat{K}=A^{t}Y\) (the normal equation), it will be a solution to the LS approach. In addition, when \(A^{t}A\) is invertible and well-conditioned, then \(\hat{K}=(A^{t}A)^{-1} A^{t}Y\) is again the unique generalized inverse solution [79].
Bayesian Estimation Approach
An alternative approach is to use Bayesian inference, which allows to find the exact parameter estimations, not only approximations. To this end, the parameters are considered as random variables, with prior distribution using prior information, e.g., from earlier data [85, 86]. Here, we need prior distributions for errors and unknown parameters.
The following equation (Eq. (5)) proposed by [85,86,87,88] is considered to estimate the pharmacokinetic parameters. The problem is to estimate the positive-vector K (the pixel intensities in an object) under a measured vector Y (e.g., a degraded image or the projections of an object) and a linear transformation A that links both vectors via
Subject to p(K), p(Y|K) and p(Y), the posterior probability distribution of K condition to Y, p(K|Y), using Bayes: rule will be [89]:
The Bayesian estimator \(\hat{K}\) can be determined by maximizing p(K|Y), such that in Eq. (10), p(Y) has no dependence on K, p(Y|K) is related to noise, and p(K) is a prior distribution of K.
The PDFs p(K) and p(Y|K) can be estimated using MET as proposed by [85, 86, 89], with the general form of the estimated model belonging to the exponential family. In the MET, initial information to define the constraints of p(K) is required to choose the model with the maximum entropy (see “Maximum Entropy Technique- Entropy as a Regularization Functional”). The posterior distribution is computed as follows
Maximum Entropy Technique—Entropy as a Regularization Functional
We propose to solve the ME problem using the regularization method. Generally, there is a unique optimizer to solve either \(J(x)={\Vert Y-Ax \Vert }^{2}+\lambda P(x)\) or
where \(\bigtriangleup _{1}\) and \(\bigtriangleup _{2}\) are two distance measures, \(\lambda\) is a regularization parameter and q is a priori solution. The important step is to choose \(\bigtriangleup _{1}\) and \(\bigtriangleup _{2}\), and determining \(\lambda\) and q. The main part of MET is maximizing Shannon’s entropy [38]:
subject to the following constraints, which are the expectations of known functions computed numerically based on the data via Taylor’s theorem [90].
in which \(\phi _{0} (x)=1\), and \(\phi _{i} (x)\), \(k=0,\ldots ,N\) are \(N+1\) known functions. The general forms of these functions are \(x^{n}\),\(\ln (x)\), \(x\ln (x)\), trigonometric or geometric functions [37]. Entropy can also be used as a regularization functional in Eq. (11). An essential challenge in this method is to specify the regularization parameter \(\lambda\). Here,
where \(D_{K-L}\) is Kullback–Leibler divergence \(D_{K-L}\) and g is an initial solution of p. J(p) is convex on \(R_{+}^{n}\) and if the solution exists, it will be unique. Using the Lagrangian technique gives the following:
with
We mentioned that p may be in nonlinear form. In the following, we briefly describe the MET regularization algorithm (MET/REG)
-
(1) Assuming \(\mu _{i}\)s as constraints which are the expected value of the known functions \(\phi (x), \in C\), computed numerically from data based on the Taylor’s theorem, \(\mu =E_{p}(\phi (X))\).
-
(2) Estimating p(x) by minimizing \(D_{K-L}\) subject to the known constraints in Eq. (13). Then, g(x) is an initial (empirical) solution for p. Using the Lagrangian, the following equation is solved
$$\begin{aligned} \begin{aligned} dp(\mu , \lambda )=exp[\lambda ^{t}[Ax]-ln Z(\lambda ) ]dg(x),\\ where~~~~ Z(\lambda )=\int _{C}exp[\lambda ^{t}[Ax]]dg(x) \end{aligned} \end{aligned}$$(17)and its parameters will be determined by \(\partial ln Z(\lambda )/ \partial \lambda _{i}\), \(i= 1, ...,M\).
-
(3) Determining the expected value of p, \(\hat{\mu }(\lambda )=E_{p}(X)=\int xdp(x, \lambda )\) as the solution of the inverse problem. The solution \(\hat{\mu }\) is a function of dual variable \(\hat{s}= A^{t}\hat{\lambda }\) by \(\hat{\mu }(s)= \bigtriangledown _{s}G(\hat{s}, q)\) in which
$$\begin{aligned} \begin{aligned}G(s, q)&= ln Z(s,q)=ln \int _{C}exp [s^{t}x]dg(x) ,\\q&=E_{g}(X)= \int _{C} xdg(x) \\ \hat{\lambda }&= argmax_{\lambda } \{ D(\lambda )= \lambda ^{t}y - G(A^{t}\hat{\lambda })\}. \end{aligned} \end{aligned}$$(18) -
(4) If the function g is a separable measure: \(g(x)=\prod _{j=1}^{N} g_{j}(x_{j})\) then p is a separable measure: \(dp(x, \lambda )=\prod _{j=1}^{N} dp_{j}(x_{j}, \lambda )\) and then,
$$\begin{aligned} G(s, q)=\sigma _{j}g_{j}(s_{j}, q_{j}), \end{aligned}$$(19)function \(g_{j}\) will be the logarithmic Laplace transform of \(g_{j}: g_{j}=ln \int exp[sx]dg_{j}(x)\).
Maximum Entropy Technique—Teaching–Learning-Based Optimization
In the previous work [76, 77], the MET/MAP and MET/TLBO have been applied to estimate the ME distribution of the AIF along with the pharmacokinetic parameters. In the following a brief description of the MET/TLBO is provided.
Applying the Lagrange multipliers approach proposed by [36, 37, 90] where Shannon’s entropy Eq. (12) is a target function with the known constraints as in Eq. (13), J(p) is
where p(x) is determined via differentiating J subject to p(x):
When setting Eq. (20) equal to zero, p(x) is as follows [38]:
and \(\lambda _{i}\) are estimated when the determined p(x) in Eq. (21) satisfies Eq. (13). In addition, determination of \(\lambda =[\lambda _{0} ,...,\lambda _{N} ]\) is the cornerstone for the specification of the family of estimated distributions. For that, TLBO is used to solve the \(N+1\) unknown parameters, as the set of \(N+1\) nonlinear equations a(\(1\le k\le m\)):
The TLBO is a commonly used technique which simulates the teaching–learning process in a class [78]. Here, a group of students are considered to be the target population, and the subjects concerning them are variables of the optimization problem. The scores of students in any subject are the value of the mentioned variables. The teacher is the best solution in the whole population and distributes his information to the students modifying the quality of learning. Additionally, the quality of a student is determined by the average value of the student’s scores in the same class. The algorithm has two main steps:
Teacher Phase
Here, the teacher attempts modifying the average scores of the students condition on their situation to produce a new result replacing the old one. This is a random step:
where C is the number of courses, \(Z_{Alt, C}\) (a vector \(1 \times C\)) is the old result with no contribution for the learners to increase their information and involves the results of every specific course, a random number \(r\in [0,1]\), \(Z_{Te, C}\) gives the most desirable solution in the entire population, \(S_{F}\) is a teaching factor ranging randomly from 1 to 2 with the same probability, and vector \(M_{C}\) ( \(1 \times C\)) is the mean scores of the class in any course. The new solution \(Z_{neu, C}\) is considered as better than the old one [91].
Learner Phase
Here, the aim is to improve the information of each student in situations in which he/she has random cooperation with other students, Eq. (24) is applied to the whole class. This way, a student is able to obtain new information from another student who has more information.
In the above, \(i=1,2\) is the solution number, \(Z_{Alt, i}\) means the lack of cooperation between \(r_{i} \in [0,1]\), and \(Z_{j}\) and \(Z_{l}\) represent two students selected randomly for \(j\ne l\) when \(Z_{j}\) provides a better objective value than \(Z_{l}\) . If the solution \(Z_{new, i}\) is better than the old one \(Z_{Alt, i}\), it is accepted.
Evaluation Procedure
All computations are done using MATLAB. To determine the general form of the ME distribution, we have applied the Kernel distribution using ‘KernelDistribution’ objects and ‘ksdensity’ in MATLAB. Regularization algorithms considered were Lasso, Ridge regression (Tikhonov regularization), and the generalized minimal residual (GMRES) method. MATLAB functions for these three methods were ‘lasso’, ‘ridge’ and ‘gmres’, respectively. The step-by-step algorithm (regularization of entropy) is provided in “Maximum Entropy Technique- Entropy as a Regularization Functional”. To assess the accuracy of each step of the algorithm, in addition to the Kullback–Leibler divergence \(D_{K-L}\), we have used symmetric measurements to evaluate the closeness of the estimated ME distribution to the empirical one. In addition, some other statistics which are robust for the comparison are mentioned:
-
(1)
Evaluation by measuring distance between the estimated PDF and the empirical one, e.g., Kullback–Leibler divergence \(D_{K-L}\)
$$\begin{aligned} D_{K-L} ({\mathop {\hat{p}}\limits ^{}} \mathrm{|}|g)=\int _{s} \hat{p}_{C_{p} } (t) \ln \frac{\hat{p}_{C_{p} } (t)}{g(C_{p} )} dt. \end{aligned}$$(26) -
(2)
Evaluation by using measures comparing the estimated values to the sample data. With the predicted values \(\hat{y}_{1} ,...,\hat{y}_{l}\) and the observed values \(y_{1} ,...,y_{l}\):
$$\begin{aligned} R-MSE=\Big [\frac{1}{l} \sum _{i=1}^{l}(y_{i} -\hat{y}_{i} )^{2} \Big ]^{1/2} , \end{aligned}$$(27)$$\begin{aligned} Chi-Square =\frac{\sum _{i=1}^{l}(y_{i} -\hat{y}_{i} )^{2} }{l-n} , \end{aligned}$$(28)$$\begin{aligned} R^{2} =1-\frac{\sum _{i=1}^{N}(y_{i} -\hat{y}_{i} )^{2} }{\sum _{i=1}^{N}(y_{i} -\bar{y})^{2} } , \end{aligned}$$(29)
Numerical Experiment
Data Description
In this study, DCE-MRI images of twelve patients before treatment were used. For each patient, once Gadolinium-DTPA was administered as the CA, 46 scans were taken at intervals of 11.9 seconds. To calculate the values of \(T_{1}\), based on calibration curves reported in [92, 93], a two-point measurement was employed. \(T_{1}\) in DCE-MRI gives the relaxation time, which measures the recovering rate of the net magnetization vector. The value of \(T_{1}\) is calculated as a ratio of a \(T_{1}\)-weighted fast low-angle shot (FLASH) image and a proton-density-weighted FLASH image. The CA concentration \(C_{T} (t)\) is measured by converting the signal intensity into \(T_{1}\) using \(T_{1}\)-weighted and proton-density-weighted images as well as data from calibration phantoms knowing \(T_{1}\) [94]. The concentration of Gd-DTPA is calculated by \(C_{T} (t)=\frac{1}{r_{1} } \left[ \frac{1}{T_{1} (t)} -\frac{1}{T_{10} } \right] ,\) where \(T_{10}\) is the \(T_{1}\) with no contrast, calculated as the average of the first four images, and \(r_{1} =4.24{} \mathrm{l/s/mmol}\) gives the in vivo longitudinal relativity of protons from Gd-DTPA. For the T1-weighted FLASH images, the obtained parameters are TR = 11 ms, TE = 4.7 ms, \(\alpha\) = 35, with the corresponding parameters for the proton-density-weighted images are TR = 350 ms, TE = 4.7 ms, \(\alpha\) = 6. All the scans had the same field of view, namely \(260\times 260\times 8\) mm per slice, making the voxel dimensions of \(1.016\times 1.0168\) mm. Each scan included three successive slices with \(256\times 256\) voxels and one slice put in the contra-lateral breast as control, which was not used for this analysis. Following the fourth scan, Gd-DTPA was injected at D = 0.1 mmol per kg body-weight using a power injector with 4 mL/s with a 20 mL saline flush also at 4 mL/s.
Example Description
We compare the proposed MET/REG algorithm with the previously proposed MET/MAP and MET/TLBO algorithms. In previous work, the Weibull distribution as PDF for the AIF turned out to satisfy most of the conditions. See Table 1 for more information about the model and estimation approaches. The PDF of the Weibull distribution with two parameters is as follows:
where \(\alpha\) and \(\beta\) are the shape and scale parameters, respectively [95, 96]. The MET tests and utilizes different moment constraints [37] and selects the minimum number of them to generate a proper PDF of the observation. The moment constraints as known functions are based on the data, and their expectations are obtained numerically via the Taylor’s theorem using the observations [90]. Note that adding more constraints does not guarantee a better ME model. The estimated PDF of the data with the initial conditions (\(\phi _{0}=1\), \(\phi _{1}=\ln (C_{p}(t))\) and \(\phi _{2}=C_{p}(t)\)) and expected values (\(1, -0.446\) and 0.335) from Taylor’s theorem fits well with the Weibull and with the family of Gamma distributions based on MET/TLBO and MET/REG (Fig. 1), respectively. For data \(\vec{C}_{p} (t)\) both models are as follows:
in which \(\lambda _{0}=-\ln (\alpha \setminus {\beta ^{\alpha }})\), \(\lambda _{1}=-(\alpha -1)\) and \(\lambda _{2}=\beta ^{-\alpha }\). Resulting values for \(\alpha\) and \(\beta\) can be found in Table 2.
The general form of the Gamma distribution and its ME model are:
in which \(\lambda _{0}=-\ln (1 \setminus \Gamma (\alpha ){\beta ^{\alpha }})\), \(\lambda _{1}=-(\alpha -1)\) and \(\lambda _{2}=\beta ^{-1}\). For Erlang distribution,
in which \(\lambda _{0}=-\ln (1 \setminus (\alpha -1)!{\beta ^{\alpha }})\), \(\lambda _{1}=-(\alpha -1)\) and \(\lambda _{2}=\beta ^{-1}\). Based on Eqs. (30) to (33), the estimated parameters for all models are presented in Table 2, respectively.
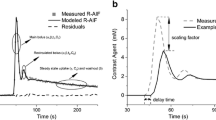
Table 3 shows Kullback–Leibler distance \(D_{K-L}\) and the entropy of different estimated AIFs for model evaluation. Higher values of entropy indicate better AIF estimation. The value of \(D_{K-L}\) is the distance between the estimated AIF and the empirical one. Lower values means the two models are close together. Figures 2 and 3 show the curve of CDFs from all proposed approaches and the empirical CDF for visual validation. In Table 4, the results are evaluated via RMSE, the goodness of fit (\(\chi ^2\)), and determination coefficient (\(R^{2}\)).
Tables 3 to 4 show that MET/TLBO and MET/REG perform the best. The proposed novel MET/REG has the lowest \(D_{K-L}\) divergence and high entropy.
For all 12 patients, similar results in regard to model performance are achieved, see Fig. 4). Nevertheless, to obtain a proper estimation of the pharmacokinetic parameters, correct estimation of the AIF is of particular importance. The measured \(D_{K-L}\) values are in the range (0, 0.1) for all 12 patients. In Fig. 5, \(K_{a}\) estimations are provided based on MET/REG and assumed AIF/ML & MET/MAP and MET/TLBO for all the patients. Most importantly, using the estimated AIF via MET/REG led to more realistic k values compared to assumed AIF, see Fig. 5 and Table 5.
Discussion and Conclusions
In this paper we investigated the application of MET and regularization functionals with some probabilistic models to solve the problem of AIF determination in DCE-MRI, a linear ill-posed inverse problem. Reasons for applying the MET in combination with the selected optimization or regularization methods to estimate the AIF, instead of using the assumed AIF, were discussed. In addition, the effect of the estimated AIF on the determination of pharmacokinetic parameters was examined.
We have shown how all these different frameworks converge to solve the linear ill-posed inverse problems. The results show that the Bayesian framework provides more tools to infer the uncertainties of the computed solutions, account for more specific knowledge of the noise and errors, estimate the hyper-parameters, and handle myopic and blind inversion problems. For that, regularization, MET, and the Bayesian estimation approach were discussed briefly. Finally, we presented some numerical results to illustrate the efficiency of the presented method. The main objective of these numerical experiments was to demonstrate the effect of different choices for prior laws or, equivalently, regularization functionals on the result. To determine the ME solution via entropy regularization, it was assumed that the existing data are represented by generalized moments, including the power and the fractional ones as a subset. However, as mentioned in the paper, the solution of an inverse problem generally depends on our prior hypothesis regarding AIF, errors and K.
The proposed MET/REG algorithm has multiple notable features: (1) applicability to distributions with any type of support, (2) efficiency in terms of computation since the ME solution is derived simply as a set of linear equations, (3) proper bias-variance, and (4) proper estimation of the distribution tails when the sample sizes are small. Given the important role of the AIF when analyzing DCE-MRI images, when determining the AIF in the image is not possible, a standard approach is to use assumed AIFs proposed in the literature. This research provides an alternative method for the assessment of the AIF from the available information. Based on the results, the estimated model using MET/REG fits well to the data and properly estimates the pharmacokinetic parameters.
Availability of Data and Material
There would be possible by personal request. Breast cancer data set provided by Paul Strickland Scanner Centre at Mount Vernon Hospital, Northwood, UK Scanner Centre at Mount Vernon Hospital, Northwood, UK
Code Availability
There would be possible by personal request.
References
Khalifa F, Soliman A, El-Baz A, Abou El-Ghar M, El-Diasty T, Gimel’farb G, et al. Models and methods for analyzing DCE-MRI: A review. Medical Physics. 2014;41(12):124301.
Fennessy FM, McKay RR, Beard CJ, Taplin ME, Tempany CM. Dynamic contrast-enhanced magnetic resonance imaging in prostate cancer clinical trials: potential roles and possible pitfalls. Translational Oncology. 2014;7(1):120–129.
Huang W, Li X, Chen Y, Li X, Chang MC, Oborski MJ, et al. Variations of dynamic contrast-enhanced magnetic resonance imaging in evaluation of breast cancer therapy response: a multicenter data analysis challenge. Translational Oncology. 2014;7(1):153.
Sobhani F, Xu C, Murano E, Pan L, Rastegar N, Kamel IR. Hypo-vascular liver metastases treated with transarterial chemoembolization: assessment of early response by volumetric contrast-enhanced and diffusion-weighted magnetic resonance imaging. Translational Oncology. 2016;9(4):287–294.
Usuda K, Iwai S, Funasaki A, Sekimura A, Motono N, Matoba M, et al. Diffusion-weighted magnetic resonance imaging is useful for the response evaluation of chemotherapy and/or radiotherapy to recurrent lesions of lung cancer. Translational Oncology. 2019;12(5):699–704.
Stoyanova R, Huang K, Sandler K, Cho H, Carlin S, Zanzonico PB, et al. Mapping tumor hypoxia in vivo using pattern recognition of dynamic contrast-enhanced MRI data. Translational Oncology. 2012;5(6):437.
Schmid VJ, Whitcher B, Padhani AR, Taylor NJ, Yang GZ. Bayesian methods for pharmacokinetic models in dynamic contrast-enhanced magnetic resonance imaging. IEEE Transactions on Medical Imaging. 2006;25(12):1627–36. Available from: http://www.ncbi.nlm.nih.gov/pubmed/17167997.
Tofts PS, Kermode AG. Measurement of the blood-brain barrier permeability and leakage space using dynamic MR imaging - 1. Fundamental concepts. Magnetic Resonance in Medicine. 1991;17(2):357–367.
Brix G, Kiessling F, Lucht R, Darai S, Wasser K, Delorme S, et al. Microcirculation and microvasculature in breast tumors: pharmacokinetic analysis of dynamic MR image series. Magnetic Resonance in Medicine. 2004;52(2):420–429.
Berg B, Stucht D, Janiga G, Beuing O, Speck O, Thovenin D. Cerebral Blood Flow in a Healthy Circle of Willis and Two Intracranial Aneurysms: Computational Fluid Dynamics Versus Four-Dimensional Phase-Contrast Magnetic Resonance Imaging. ASME J Biomech Eng. 2014;15:041003. https://doi.org/10.1115/1.4026108.
Orton MR, Collins DJ, Walker-Samuel S, d’Arcy JA, Hawkes DJ, Atkinson D, et al. Bayesian estimation of pharmacokinetic parameters for DCE-MRI with a robust treatment of enhancement onset time. Phys Med Biol. 2007;52:2393–2408.
Dikaios N, Arridge S, Hamy V, Punwani S, Atkinson D. Direct parametric reconstruction from undersampled (k, t)-space data in dynamic contrast enhanced MRI. Medical Image Analysis. 2014;18(7):989–1001.
Bender R, Heinemann L. Fitting nonlinear regression models with correlated errors to individual pharmacodynamic data using SAS software. Journal of Pharmacokinetics and Biopharmaceutics. 1995;23(1):87–100.
Cheng HLM. T1 measurement of flowing blood and arterial input function determination for quantitative 3D T1-weighted DCE-MRI. Journal of Magnetic Resonance Imaging : JMRI. 2007;25(5):1073–8.
Gauthier M. Impact of the arterial input function on microvascularization parameter measurements using dynamic contrast-enhanced ultrasonography. World Journal of Radiology. 2012;4(7):291.
Larsson HBW, Tofts PS. Measurement of blood-brain barrier permeability using dynamic Gd-DTPA scanning –a comparison of methods. Magnetic Resonance in Medicine. 1992;24(1):174–176.
Cheng HLM. Investigation and optimization of parameter accuracy in dynamic contrast-enhanced MRI. Journal of Magnetic Resonance Imaging. 2008;28(3):736–743. Available from: http://dx.doi.org/10.1002/jmri.21489.
Lavini C. Simulating the effect of input errors on the accuracy of Tofts’ pharmacokinetic model parameters. Magnetic Resonance Imaging. 2015;33(2):222–235.
Peled S, Vangel M, Kikinis R, Tempany CM, Fennessy FM, Fedorov A. Selection of fitting model and arterial input function for repeatability in dynamic contrast-enhanced prostate MRI. Academic Radiology. 2019;26(9):e241–e251.
Huang W, Chen Y, Fedorov A, Li X, Jajamovich GH, Malyarenko DI, et al. The impact of arterial input function determination variations on prostate dynamic contrast-enhanced magnetic resonance imaging pharmacokinetic modeling: a multicenter data analysis challenge. Tomography. 2016;2(1):56–66.
Huang W, Chen Y, Fedorov A, Li X, Jajamovich GH, Malyarenko DI, et al. The impact of arterial input function determination variations on prostate dynamic contrast-enhanced magnetic resonance imaging pharmacokinetic modeling: a multicenter data analysis challenge, part II. Tomography. 2019;5(1):99–109.
Keil VC, Mädler B, Gieseke J, Fimmers R, Hattingen E, Schild HH, et al. Effects of arterial input function selection on kinetic parameters in brain dynamic contrast-enhanced MRI. Magnetic resonance imaging. 2017;40:83–90.
Weinmann HJ, Laniado M, Mützel W. Pharmokinetics of Gd-DTPA/Dimeglumine after intravenous injection into healthy volunteers. Physiological Chemistry & Physics & Medical NMR. 1984;16:167–172.
Fritz-Hansen T, Rostrup E, Larsson HBW, Sø ndergaard L, Ring P, Henriksen O. Measurement of the Arterial Concentration of Gd-DTPA Using MRI: A step toward Quantitative Perfusion Imaging. Magnetic Resonance in Medicine. 1996;36(2):225–231.
Parker GJ, Roberts C, Macdonald A, Buonaccorsi GA, Cheung S, Buckley DL, et al. Experimentally-derived functional form for a population-averaged high-temporal-resolution arterial input function for dynamic contrast-enhanced MRI. Magnetic Resonance in Medicine: An Official Journal of the International Society for Magnetic Resonance in Medicine. 2006;56(5):993–1000.
Rata M, Collins DJ, Darcy J, Messiou C, Tunariu N, Desouza N, et al. Assessment of repeatability and treatment response in early phase clinical trials using DCE-MRI: comparison of parametric analysis using MR-and CT-derived arterial input functions. European radiology. 2016;26(7):1991–1998.
Rijpkema M, Kaanders JH, Joosten FB, van der Kogel AJ, Heerschap A. Method for quantitative mapping of dynamic MRI contrast agent uptake in human tumors. Journal of Magnetic Resonance Imaging: An Official Journal of the International Society for Magnetic Resonance in Medicine. 2001;14(4):457–463.
Ashton E, Raunig D, Ng C, Kelcz F, McShane T, Evelhoch J. Scan-rescan variability in perfusion assessment of tumors in MRI using both model and data-derived arterial input functions. Journal of Magnetic Resonance Imaging: An Official Journal of the International Society for Magnetic Resonance in Medicine. 2008;28(3):791–796.
Shao J, Zhang Z, Liu H, Song Y, Yan Z, Wang X, et al. DCE-MRI pharmacokinetic parameter maps for cervical carcinoma prediction. Computers in Biology and Medicine. 2020;118:103634.
Lingala SG, Guo Y, Bliesener Y, Zhu Y, Lebel RM, Law M, et al. Tracer kinetic models as temporal constraints during brain tumor DCE-MRI reconstruction. Medical physics. 2020;47(1):37–51.
Zou J, Balter JM, Cao Y. Estimation of pharmacokinetic parameters from DCE-MRI by extracting long and short time-dependent features using an LSTM network. Medical Physics. 2020;47(8):3447–3457.
Dikaios N. Stochastic Gradient Langevin dynamics for joint parameterization of tracer kinetic models, input functions, and T1 relaxation-times from undersampled k-space DCE-MRI. Medical Image Analysis. 2020;62:101690.
Fan S, Bian Y, Wang E, Kang Y, Wang DJ, Yang Q, et al. An automatic estimation of arterial input function based on multi-stream 3D CNN. Frontiers in Neuroinformatics. 2019;p. 49.
Reishofer G, Bammer R, Moseley M, Stollberger R. Automatic arterial input function detection from dynamic contrast enhanced MRI data. In: 11th Scientific Meeting of the International Society for Magnetic Resonance in Medicine, Toronto, Ontario, Canada; 2003.
Jaynes ET. Information Theory and Statistical Mechanics. Physics Reveiw. 1957;106(4):620–630.
Pougaza DB, Djafari AM. Maximum Entropy Copulas. AIP Conference Proceeding. 2011;p. 2069–2072.
Ebrahimi N, Soofi ES, Soyer R. Multivariate maximum entropy identification, transformation, and dependence. Multi Analys. 2008;99:1217–1231.
Thomas A, Cover TM. Elements of Information Theory. New Jersey: John Wiley; 2006.
Cofré R, Herzog R, Corcoran D, Rosas FE. A comparison of the maximum entropy principle across biological spatial scales. Entropy. 2019;21(10):1009.
Jaynes ET. Probability theory: The logic of science. Cambridge university press; 2003.
Pitera JW, Chodera JD. On the use of experimental observations to bias simulated ensembles. Journal of Chemical Theory and Computation. 2012;8(10):3445–3451.
Hopf TA, Colwell LJ, Sheridan R, Rost B, Sander C, Marks DS. Three-dimensional structures of membrane proteins from genomic sequencing. Cell. 2012;149(7):1607–1621.
Roux B, Weare J. On the statistical equivalence of restrained-ensemble simulations with the maximum entropy method. The Journal of Chemical Physics. 2013;138(8):02B616.
Cavalli A, Camilloni C, Vendruscolo M. Molecular dynamics simulations with replica-averaged structural restraints generate structural ensembles according to the maximum entropy principle. The Journal of Chemical Physics. 2013;138(9):03B603.
Jennings RC, Belgio E, Zucchelli G. Does maximal entropy production play a role in the evolution of biological complexity? A biological point of view. Rendiconti Lincei Scienze Fisiche e Naturali. 2020;p. 1–10.
Ekeberg M, Lövkvist C, Lan Y, Weigt M, Aurell E. Improved contact prediction in proteins: using pseudolikelihoods to infer Potts models. Physical Review E. 2013;87(1):012707.
Boomsma W, Ferkinghoff-Borg J, Lindorff-Larsen K. Combining experiments and simulations using the maximum entropy principle. PLoS Comput Biol. 2014;10(2):e1003406.
Zhang B, Wolynes PG. Topology, structures, and energy landscapes of human chromosomes. Proceedings of the National Academy of Sciences. 2015;112(19):6062–6067.
Cesari A, Reißer S, Bussi G. Using the maximum entropy principle to combine simulations and solution experiments. Computation. 2018;6(1):15.
Farré P, Emberly E. A maximum-entropy model for predicting chromatin contacts. PLoS Computational Biology. 2018;14(2):e1005956.
Remacle F, Kravchenko-Balasha N, Levitzki A, Levine RD. Information-theoretic analysis of phenotype changes in early stages of carcinogenesis. Proceedings of the National Academy of Sciences. 2010;107(22):10324–10329.
Sanguinetti G, et al. Gene regulatory network inference: an introductory survey. In: Gene Regulatory Networks. Springer; 2019. p. 1–23.
Locasale JW, Wolf-Yadlin A. Maximum entropy reconstructions of dynamic signaling networks from quantitative proteomics data. PloS One. 2009;4(8):e6522.
Graeber T, Heath J, Skaggs B, Phelps M, Remacle F, Levine RD. Maximal entropy inference of oncogenicity from phosphorylation signaling. Proceedings of the National Academy of Sciences. 2010;107(13):6112–6117.
Sharan R, Karp RM. Reconstructing Boolean models of signaling. Journal of Computational Biology. 2013;20(3):249–257.
Quadeer AA, McKay MR, Barton JP, Louie RH. MPF-BML: a standalone GUI-based package for maximum entropy model inference. Bioinformatics. 2020;36(7):2278–2279.
Tkačik G, Prentice JS, Balasubramanian V, Schneidman E. Optimal population coding by noisy spiking neurons. Proceedings of the National Academy of Sciences. 2010;107(32):14419–14424.
Ohiorhenuan IE, Mechler F, Purpura KP, Schmid AM, Hu Q, Victor JD. Sparse coding and high-order correlations in fine-scale cortical networks. Nature. 2010;466(7306):617–621.
Yeh FC, Tang A, Hobbs JP, Hottowy P, Dabrowski W, Sher A, et al. Maximum entropy approaches to living neural networks. Entropy. 2010;12(1):89–106.
Granot-Atedgi E, Tkačik G, Segev R, Schneidman E. Stimulus-dependent maximum entropy models of neural population codes. PLoS Comput Biol. 2013;9(3):e1002922.
Tkačik G, Marre O, Mora T, Amodei D, Berry II MJ, Bialek W. The simplest maximum entropy model for collective behavior in a neural network. Journal of Statistical Mechanics: Theory and Experiment. 2013;2013(03):P03011.
Ferrari U, Obuchi T, Mora T. Random versus maximum entropy models of neural population activity. Physical Review E. 2017;95(4):042321.
Rostami V, Mana PP, Grün S, Helias M. Bistability, non-ergodicity, and inhibition in pairwise maximum-entropy models. PLoS Computational Biology. 2017;13(10):e1005762.
Nghiem TA, Teleńczuk B, Marre O, Destexhe A, Ferrari U. Maximum entropy models reveal the correlation structure in cortical neural activity during wakefulness and sleep. bioRxiv. 2018;p. 243857.
Mora T, Walczak AM, Bialek W, Callan CG. Maximum entropy models for antibody diversity. Proceedings of the National Academy of Sciences. 2010;107(12):5405–5410.
Santolini M, Mora T, Hakim V. A general pairwise interaction model provides an accurate description of in vivo transcription factor binding sites. PloS One. 2014;9(6):e99015.
Fariselli P, Taccioli C, Pagani L, Maritan A. DNA sequence symmetries from randomness: the origin of the Chargaff’s second parity rule. Briefings in Bioinformatics. 2020.
Fernandez-de Cossio-Diaz J, Mulet R. Maximum entropy and population heterogeneity in continuous cell cultures. PLoS Computational Biology. 2019;15(2):e1006823.
Thapliyal M, Ahuja NJ, Shankar A, Cheng X, Kumar M. A differentiated learning environment in domain model for learning disabled learners. Journal of Computing in Higher Education. 2021;p. 1–23.
Kumar S, Viral R, Deep V, Sharma P, Kumar M, Mahmud M, et al. Forecasting major impacts of COVID-19 pandemic on country-driven sectors: challenges, lessons, and future roadmap. Personal and Ubiquitous Computing. 2021;p. 1–24.
Dhasarathan C, Kumar M, Srivastava AK, Al-Turjman F, Shankar A, Kumar M. A bio-inspired privacy-preserving framework for healthcare systems. The Journal of Supercomputing. 2021;77(10):11099–11134.
Jackson A, Constable C, Gillet N. Maximum entropy regularization of the geomagnetic core field inverse problem. Geophysical Journal International. 2007;171(3):995–1004.
De Martino A, De Martino D. An introduction to the maximum entropy approach and its application to inference problems in biology. Heliyon. 2018;4(4):e00596.
Chakradar M, Aggarwal A, Cheng X, Rani A, Kumar M, Shankar A. A Non-invasive Approach to Identify Insulin Resistance with Triglycerides and HDL-c Ratio Using Machine learning. Neural Processing Letters. 2021;p. 1–21.
Aggarwal A, Alshehri M, Kumar M, Sharma P, Alfarraj O, Deep V. Principal component analysis, hidden Markov model, and artificial neural network inspired techniques to recognize faces. Concurrency and Computation: Practice and Experience. 2021;33(9):e6157.
Farsani ZA, Schmid VJ. Maximum Entropy Approach in Dynamic Contrast-Enhanced Magnetic Resonance Imaging. MEthods of Information in Medicine. 2017;56(06):461–468.
Amini Farsani Z, Schmid VJ. Modified Maximum Entropy Method and Estimating the AIF via DCE-MRI Data Analysis. Entropy. 2022;24(2):155.
Rao RV, Savsani VJ, Vakharia D. Teaching-learning-based optimization: a novel method for constrained mechanical design optimization problems. Computer-Aided Design. 2011;43(3):303–315.
Mohammad-Djafari A, Giovannelli JF, Demoment G, Idier J. Regularization, maximum entropy and probabilistic methods in mass spectrometry data processing problems. International Journal of Mass Spectrometry. 2002;215(1-3):175–193.
Leach MO, Brindle KM, Evelhoch JL, Griffiths JR, Horsman MR, Jackson A, et al. The assessment of antiangiogenic and antivascular therapies in early-stage clinical trials using magnetic resonance imaging: issues and recommendations. British Journal of Cancer. 2005;92:1599–1610.
Choyke PL, Dwyer AJ, Knopp MV. Functional tumor imaging withdynamic contrast-enhanced magnetic resonance imaging. Magnetic Resonance in Medicine. 2003;17:509–520.
Murase K. Efficient method for calculating kinetic parameters using T1-weighted dynamic contrast-enhanced magnetic resonance imaging. MAgnetic Resonance in Medicine. 2004;51(4):858–862.
Mohammad-Djafari A. Bayesian Image Processing. In: Fifth Int. Conf. on Modelling, Computation and Optimization in Information Systems and Management Sciences (MCO 2004), Metz, France; 2004. .
Bertsekas D. Nonlinear Programming, vol. 43. Belmont, MA, USA: Athena Scientific. 1995;.
Mohammad-Djafari A, Demoment G. Estimating priors in maximum entropy image processing. In: International Conference on Acoustics, Speech, and Signal Processing. IEEE; 1990. p. 2069–2072.
Mohammad-Djafari A. A full Bayesian approach for inverse problems. In: Maximum entropy and Bayesian methods. Springer; 1996. p. 135–144.
Denisova N. Bayesian maximum-a-posteriori approach with global and local regularization to image reconstruction problem in medical emission tomography. Entropy. 2019;21(11):1108.
Sparavigna AC. Entropy in image analysis. Multidisciplinary Digital Publishing Institute; 2019.
Elfving T. An Algorithm for Maximum Entropy Image Reconstruction form Noisy Data. MathlcomputModeling. 1989;12:729–745.
Casella G, Berger RL. statistical inference 2. CA, USA: Duxbury; 2002.
García JAM, Mena AJG. Optimal distributed generation location and size using a modified teaching–learning based optimization algorithm. International Journal of Electrical Power & Energy Systems. 2013;50:65–75.
Parker GJ, Suckling J, Tanner SF, Padhani AR, Revell PB, Husband JE, et al. Probing tumor microvascularity by measurement, analysis and display of contrast agent uptake kinetics. Journal of Magnetic Resonance Imaging. 1997;7(3):564–574.
d’Arcy JA, Collins DJ, Padhani AR, Walker-Samuel S, Suckling J, Leach MO. Magnetic resonance imaging workbench: analysis and visualization of dynamic contrast-enhanced MR imaging data. Radiographics. 2006;26(2):621–632.
Buckley DL, Parker GJM. Measuring Contrast Agent Concentration in \(T_1\)-Weighted Dynamic Contrast-Enhanced MRI. In: Jackson A, Parker GJM, Buckley DL, editors. Dynamic Contrast-Enhanced Magntic Resoncance Imaging in Oncology. Berlin, Heidelberg, New York: Springer; 2005. p. 69–80.
Bain LJ, Antle CE. Estimation of parameters in the weibdl distribution. Technometrics. 1967;9(4):621–627.
Stevens M, Smulders P. The estimation of the parameters of the Weibull wind speed distribution for wind energy utilization purposes. Wind Engineering. 1979;p. 132–145.
Justus C, Hargraves W, Mikhail A, Graber D. Methods for estimating wind speed frequency distributions. JoUrnal of Applied Meteorology. 1978;17(3):350–353.
Werapun W, Tirawanichakul Y, Waewsak J. Comparative study of five methods to estimate Weibull parameters for wind speed on Phangan Island, Thailand. Energy Procedia. 2015;79:976–981.
Zhang H, Yu YJ, Liu ZY. Study on the Maximum Entropy Principle applied to the annual wind speed probability distribution: A case study for observations of intertidal zone anemometer towers of Rudong in East China Sea. Applied Energy. 2014;114:931–938.
Funding
Open Access funding enabled and organized by Projekt DEAL.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Consent for Publication
I give my consent for the publication of identifiable details, which can include photograph(s) and/or videos and/or case history and/or details within the text (“Material”) to be published in the above Journal and Article.
Conflicts of Interest
The authors whose names are listed immediately below certify that they have NO affiliations with or involvement in any organization or entity with any financial interest (such as honoraria; educational grants; participation in speakers’ bureaus; membership, employment, consultancies, stock ownership, or other equity interest; and expert testimony or patent-licensing arrangements), or non-financial Title Page interest (such as personal or professional relationships, affiliations, knowledge or beliefs) in the subject matter or materials discussed in this manuscript.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Amini Farsani, Z., Schmid, V.J. Maximum Entropy Technique and Regularization Functional for Determining the Pharmacokinetic Parameters in DCE-MRI. J Digit Imaging 35, 1176–1188 (2022). https://doi.org/10.1007/s10278-022-00646-3
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10278-022-00646-3