Abstract
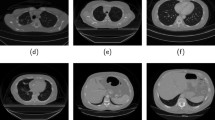
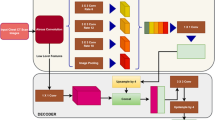
A robust lung segmentation method using a deep convolutional neural network (CNN) was developed and evaluated on high-resolution computed tomography (HRCT) and volumetric CT of various types of diffuse interstitial lung disease (DILD). Chest CT images of 617 patients with various types of DILD, including cryptogenic organizing pneumonia (COP), usual interstitial pneumonia (UIP), and nonspecific interstitial pneumonia (NSIP), were scanned using HRCT (1–2-mm slices, 5–10-mm intervals) and volumetric CT (sub-millimeter thickness without intervals). Each scan was segmented using a conventional image processing method and then manually corrected by an expert thoracic radiologist to create gold standards. The lung regions in the HRCT images were then segmented using a two-dimensional U-Net architecture with the deep CNN, using separate training, validation, and test sets. In addition, 30 independent volumetric CT images of UIP patients were used to further evaluate the model. The segmentation results for both conventional and deep-learning methods were compared quantitatively with the gold standards using four accuracy metrics: the Dice similarity coefficient (DSC), Jaccard similarity coefficient (JSC), mean surface distance (MSD), and Hausdorff surface distance (HSD). The mean and standard deviation values of those metrics for the HRCT images were 98.84 ± 0.55%, 97.79 ± 1.07%, 0.27 ± 0.18 mm, and 25.47 ± 13.63 mm, respectively. Our deep-learning method showed significantly better segmentation performance (p < 0.001), and its segmentation accuracies for volumetric CT were similar to those for HRCT. We have developed an accurate and robust U-Net-based DILD lung segmentation method that can be used for patients scanned with different clinical protocols, including HRCT and volumetric CT.





Similar content being viewed by others
References
Franks TJ, Galvin JR, Frazier AA: The use and impact of HRCT in diffuse lung disease. Current Diagnostic Pathology. 10(4):279–290, 2004
Massoptier L, Misra A, Sowmya A, Casciaro S: Combining Graph-Cut Technique and Anatomical Knowledge for Automatic Segmentation of Lungs Affected By Diffuse Parenchymal Disease in HRCT mages. International Journal of Image and Graphics. 11(04):509–529, 2011
Jun S, Park B, Seo JB, Lee S, Kim N: Development of a Computer-Aided Differential Diagnosis System to Distinguish Between Usual Interstitial Pneumonia and Non-specific Interstitial Pneumonia Using Texture-and Shape-Based Hierarchical Classifiers on HRCT mages. Journal of Digital Imaging 31(2):235–244, 2018
Kim GB, Jung K-H, Lee Y, Kim H-J, Kim N, Jun S, Seo JB, Lynch DA: Comparison of Shallow and Deep Learning Methods on Classifying the Regional Pattern of Diffuse Lung Disease. Journal of Digital Imaging 31(4):415–424, 2018
Kalinovsky A, Kovalev V. Lung image segmentation using deep learning methods and convolutional neural networks. International Conference on Pattern Recognition and Information Processing (PRIP-2016), Minsk, Belarus, 2016
Krizhevsky A, Sutskever I, Hinton GE. Imagenet classification with deep convolutional neural networks. Advances in neural information processing systems, NIPS 2012, http://papers.nips.cc/paper/4824-imagenet-classification-with-deep-convolutional-neural-networks.pdf. Accessed 5 May 2018
LeCun Y, Boser B, Denker JS, Henderson D, Howard RE, Hubbard W, Jackel LD: Backpropagation applied to handwritten zip code recognition. Neural computation. 1(4):541–551, 1989
Deng J, Dong W, Socher R, Li L, Li, K, Fei-Fei L. ImageNet: A Large-Scale Hierarchical Image Database. Proc of IEEE Computer Vision and Pattern Recognition, 2009. 2009, http://www.image-net.org/papers/imagenet_cvpr09.pdf. Accessed 5 May 2018
Krizhevsky A, Hinton G. Learning multiple layers of features from tiny images. Citeseer, 2009
Schmidhuber J: Deep learning in neural networks: An overview. Neural networks. 61:85–117, 2015
Raghu G, Collard HR, Egan JJ, Martinez FJ, Behr J, Brown KK, Colby TV, Cordier JF, Flaherty KR, Lasky JA, Lynch DA, Ryu JH, Swigris JJ, Wells AU, Ancochea J, Bouros D, Carvalho C, Costabel U, Ebina M, Hansell DM, Johkoh T, Kim DS, King te Jr, Kondoh Y, Myers J, Müller NL, Nicholson AG, Richeldi L, Selman M, Dudden RF, Griss BS, Protzko SL, Schünemann HJ, ATS/ERS/JRS/ALAT Committee on Idiopathic Pulmonary Fibrosis: An official ATS/ERS/JRS/ALAT statement: idiopathic pulmonary fibrosis: evidence-based guidelines for diagnosis and management. American Journal of Respiratory and Critical Care Medicine 183(6):788–824, 2011
McCormick M, Johnson H, Ibanez L. The ITK Software Guide: The insight segmentation and registration toolkit, 2015, https://itk.org/. Accessed 5 May 2018
Armato SG, Giger ML, Moran CJ, Blackburn JT, Doi K, MacMahon H: Computerized detection of pulmonary nodules on CT scans. Radiographics 19(5):1303–1311, 1999
Hu S, Hoffman EA, Reinhardt JM: Automatic lung segmentation for accurate quantitation of volumetric X-ray CT images. IEEE Transactions on Medical Imaging 20(6):490–498, 2001
Ronneberger O, Fischer P, Brox T. U-net: Convolutional networks for biomedical image segmentation. arXiv preprint arXiv:1505.04597, 2015
Long J, Shelhamer E, Darrell T, editors. Fully convolutional networks for semantic segmentation. IEEE Transactions on Pattern Analysis and Machine Intelligence Archive 39(4)640–651, 2017
Çiçek Ö, Abdulkadir A, Lienkamp SS, Brox T, Ronneberger O, editors. 3D U-Net: learning dense volumetric segmentation from sparse annotation. International Conference on Medical Image Computing and Computer-Assisted Intervention: Springer, 2016
Milletari F, Navab N, Ahmadi S-A, editors. V-net: Fully convolutional neural networks for volumetric medical image segmentation. 3D Vision (3DV), 2016 Fourth International Conference on; IEEE, 2016
Kingma DP, Ba J: Adam: A method for stochastic optimization. arXiv preprint arXiv:14126980. 2014.
Van Ginneken B, Heimann T, Styner M. 3D segmentation in the clinic: A grand challenge. 3D segmentation in the clinic: a grand challenge. 7–15, 2007, http://sliver07.org/p7.pdf. Accessed 5 May 2018
Jones E, Oliphant T, Peterson P. SciPy: Open source scientific tools for Python. 2001, http://www.scipy.org/. Accessed 5 May 2018
Funding
This work was supported by the Industrial Strategic technology development program (10072064) funded by the Ministry of Trade Industry and Energy (MI, Korea) and by Kakao and Kakao Brain corporations.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of Interest
The authors declare that they have no conflicts of interest.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Park, B., Park, H., Lee, S.M. et al. Lung Segmentation on HRCT and Volumetric CT for Diffuse Interstitial Lung Disease Using Deep Convolutional Neural Networks. J Digit Imaging 32, 1019–1026 (2019). https://doi.org/10.1007/s10278-019-00254-8
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10278-019-00254-8