Abstract
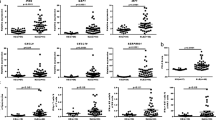
Systemic lupus erythematosus (SLE) is a multisystem autoimmune disease with complex genetic predisposing factors involved. PU.1 is an important member of the ETS transcription factors family which has diverse functions such as regulating the proliferation, differentiation of immune cells and multiple inflammatory cytokines. Previous studies preliminary explored the relation between PU.1 and SLE. To further explain the potential role of PU.1 in the pathogenesis of SLE, 40 SLE patients and 20 age-sex matched healthy controls (HC) were recruited in this study. Flow cytometry was used to test the percentages of CD4+PU.1+T cells in peripheral blood mononuclear cells (PBMCs) from patients with SLE and HC. Expression levels of PU.1 mRNA in CD4+T cells from SLE patients and HC were analyzed by real-time transcription-polymerase chain reaction. Expression levels of plasma IL-1β, IL-9, IL-18, IL-6, IFN-α, TNF-α, IL-10 and TGF-β1 were measured by enzyme-linked immunosorbent assay. The percentage of CD4+PU.1+T cells in PBMCs from patients with SLE was significantly higher than that from HC (P < 0.001). In addition, the PU.1 mRNA expression in CD4+T cells from SLE patients was increased than that from HC (P = 0.002). In SLE patients, no significant correlation was found between the percentage of CD4+PU.1+T cells and the expression of PU.1 mRNA in CD4+T cells (P > 0.05). Associations of PU.1 mRNA expression in CD4+T cells with major clinical and laboratory parameters of SLE patients were also analyzed, but no significant correlations were found. Consistent with previous studies, SLE patients had increased IL-1β, IL-18, IL-6, IFN-α, TNF-α and IL-10 plasma concentrations than HC (P < 0.01). The expression level of plasma TGF-β1 was significantly decreased in SLE patients than in HC (P < 0.001). In SLE patients, the expression level of IL-1β was positive correlated with PU.1 mRNA expression in CD4+T cells (P = 0.001). Our study first time evaluated the expression profile of PU.1 in CD4+T cells from SLE patients confirming that PU.1 may participate in the pathogenesis of SLE.







Similar content being viewed by others
Data availability
The datasets used or analyzed during the current study are available from the corresponding author on reasonable request.
Change history
05 June 2021
A Correction to this paper has been published: https://doi.org/10.1007/s10238-021-00731-x
References
Rees F, Doherty M, Grainge MJ, et al. The worldwide incidence and prevalence of systemic lupus erythematosus: a systematic review of epidemiological studies. Rheumatology. 2017;56(11):1945–61.
Zhang S, Ye Z, Li C, et al. Chinese systemic lupus erythematosus treatment and research group (CSTAR) registry XI: gender impact on long-term outcome. Lupus. 2018;28(5):635–41.
Tsokos GC. Systemic lupus erythematosus. N Engl J Med. 2011;365(22):2110–21.
Gallant S, Gilkeson G. ETS transcription factors and regulation of immunity. Arch Immunol Ther Exp (Warsz). 2006;54(3):149–63.
Moreau-Gachelin F, Tavitian A, Tambourin P. Spi-1 is a putative oncogene in virally induced murine erythroleukaemias. Nature. 1988;331(6153):277–80.
Dakic A, Wu L, Nutt SL. Is PU.1 a dosage-sensitive regulator of haemopoietic lineage commitment and leukaemogenesis? Trends Immunol. 2007;28(3):108–14.
Turkistany SA, DeKoter RP. The transcription factor PU.1 is a critical regulator of cellular communication in the immune system. Arch Immunol Ther Exp (Warsz). 2011;59(6):431–40.
Huang W, Horvath E, Eklund EA. PU.1, interferon regulatory factor (IRF) 2, and the interferon consensus sequence-binding protein (ICSBP/IRF8) cooperate to activate NF1 transcription in differentiating myeloid cells. J Biol Chem. 2007;282(9):6629–43.
Cheng J, Wu R, Long L, et al. miRNA-451 a targets IFN regulatory factor 8 for the progression of systemic lupus erythematosus. Inflammation. 2017;40(2):676–87.
Javierre BM, Fernandez AF, Richter J, et al. Changes in the pattern of DNA methylation associate with twin discordance in systemic lupus erythematosus. Genome Res. 2010;20(2):170–9.
Hikami K, Kawasaki A, Ito I, et al. Association of a functional polymorphism in the 3’-untranslated region of SPI1 with systemic lupus erythematosus. Arthritis Rheum. 2011;63(3):755–63.
Fukai T, Nishiyama C, Kanada S, et al. Involvement of PU.1 in the transcriptional regulation of TNF-alpha. Biochem Biophys Res Commun. 2009;388(1):102–6.
Larsson L, Rymo L, Berglundh T. Sp1 binds to the G allele of the-1087 polymorphism in the IL-10 promoter and promotes IL-10 mRNA transcription and protein production. Genes Immun. 2010;11(2):181–7.
Soroosh P, Doherty TA. Th9 and allergic disease. Immunology. 2009;127(4):450–8.
Ouyang H, Shi Y, Liu Z, et al. Increased interleukin-9 and CD4+IL-9+ T cells in patients with systemic lupus erythematosus. Mol Med Rep. 2013;7(3):1031–7.
Chang HC, Sehra S, Goswami R, et al. The transcription factor PU.1 is required for the development of IL-9-producing T cells and allergic inflammation. Nat Immunol. 2010;11(6):527–34.
Kim YM, Kang HS, Paik SG, et al. Roles of IFN consensus sequence binding protein and PU.1 in regulating IL-18 gene expression. J Immunol. 1999;163(4):2000–7.
Kominato Y, Galson D, Waterman WR, et al. Monocyte expression of the human prointerleukin 1 beta gene (IL1B) is dependent on promoter sequences which bind the hematopoietic transcription factor Spi-1/PU.1. Mol Cell Biol. 1995;163(4):58–68.
Marín-Rosales M, Cruz A, Salazar-Camarena DC, et al. High BAFF expression associated with active disease in systemic lupus erythematosus and relationship with rs9514828C>T polymorphism in TNFSF13B gene. Clin Exp Med. 2019;19(2):183–90.
Aboelenein HR, Hamza MT, Marzouk H, et al. Reduction of CD19 autoimmunity marker on B cells of paediatric SLE patients through repressing PU.1/TNF-α/BAFF axis pathway by miR-155. Growth Factors. 2017;35(2–3):49–60.
Hochberg MC. Updating the American College of Rheumatology revised criteria for the classification of systemic lupus erythematosus. Arthritis Rheum. 1997;40(9):1725.
Gladman DD, Iban~ez D, Urowitz MB. Systemic lupus erythematosus disease activity index 2000. J Rheumatol. 2002;29(2):288–91.
Spain LM, Guerriero A, Kunjibettu S, et al. T cell development in PU.1-deficient mice. J Immunol. 1999;163(5):2681–7.
Chang HC, Zhang S, Thieu VT, et al. PU.1 expression delineates heterogeneity in primary Th2 cells. Immunity. 2005;22(6):693–703.
Chang HC, Han L, Jabeen R, et al. PU.1 regulates TCR expression by modulating GATA-3 activity. J Immunol. 2009;183(8):4887–94.
Ghisletti S, Barozzi I, Mietton F, et al. Identification and characterization of enhancers controlling the inflammatory gene expression program in macrophages. Immunity. 2010;32(3):317–28.
Heinz S, Benner C, Spann N, et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell. 2010;38(4):576–89.
Xie Z, Qu Y, Shen P, et al. PU.1 attenuates TNF-α-induced proliferation and cytokine release of rheumatoid arthritis fibroblast-like synoviocytes by regulating miR-155 activity. Mol Med Rep. 2018;17(6):8349–56.
Umazume A, Kezuka T, Matsuda R, et al. Role of PU1 expression as an inflammatory marker in experimental autoimmune uveoretinitis. Ocul Immunol Inflamm. 2018;26(6):951–63.
Shakerian L, Ghorbani S, Talebi F, et al. MicroRNA-150 targets PU.1 and regulates macrophage differentiation and function in experimental autoimmune encephalomyelitis. J Neuroimmunol. 2018;323:167–74.
Wu Z, Zhang S, Li P, et al. Association between complement 4 copy number variation and systemic lupus erythematosus: a meta-analysis. Clin Exp Med. 2020;20(4):627–34.
Dozmorov MG, Wren JD, Alarcón-Riquelme ME. Epigenomic elements enriched in the promoters of autoimmunity susceptibility genes. Epigenetics. 2014;9(2):276–85.
Huang Y, Chen L, Zhu B, et al. Evaluation of systemic lupus erythematosus disease activity using anti-α-enolase antibody and RDW. Clin Exp Med. 2020. https://doi.org/10.1007/s10238-020-00657-w.
Veldhoen M, Uyttenhove C, van Snick J, et al. Transforming growth factor-beta ‘reprograms’ the differentiation of T helper 2 cells and promotes an interleukin 9-producing subset. Nat Immunol. 2008;9(12):1341–6.
Ouyang H, Shi Y, Liu Z, et al. Increased interleukin-9 and CD4+IL-9+T cells in patients with systemic lupus erythematosus. Mol Med Rep. 2013;7(3):1031–7.
Elyaman W, Bradshaw EM, Uyttenhove C, et al. IL-9 induces differentiation of TH17 cells and enhances function of FoxP3+ natural regulatory T cells. Proc Natl Acad Sci USA. 2009;106(31):12885–90.
Marecki S, Riendeau CJ, Liang MD, et al. PU.1 and multiple IFN regulatory factor proteins synergize to mediate transcriptional activation of the human IL-1 beta gene. J Immunol. 2001;166(11):6829–38.
Lemay S, Mao C, Singh AK. Cytokine gene expression in the MRL/lpr model of lupus nephritis. Kidney Int. 1996;50(1):85–93.
McCarthy EM, Smith S, Lee RZ, et al. The association of cytokines with disease activity and damage scores in systemic lupus erythematosus patients. Rheumatol (Oxford). 2014;53(9):1586–94.
Umare V, Pradhan V, Nadkar M, et al. Effect of proinflammatory cytokines (IL-6, TNFα, and IL-1β) on clinical manifestations in Indian SLE patients. Mediators Inflamm. 2014;2014:385297.
Mende R, Vincent FB, Kandane-Rathnayake R, et al. Analysis of serum interleukin (IL)-1β and IL-18 in systemic lupus erythematosus. Front Immunol. 2018;9:1250.
Koyama N, Hoelzer D, Ottmann OG. Regulation of human IL-18 gene expression: interaction of PU.1 with GC-box binding protein is involved in human IL-18 expression in myeloid cells. Eur J Immunol. 2004;34(3):817–26.
Sabry A, Sheashaa H, El-Husseini A, et al. Proinflammatory cytokines (TNF-alpha and IL-6) in Egyptian patients with SLE: its correlation with disease activity. Cytokine. 2006;35(3–4):148–53.
Paradowska-Gorycka A, Wajda A, Stypinska B, et al. Variety of endosomal TLRs and interferons (IFN-alpha, IFN-beta, IFN-gamma) expression profiles in patients with SLE, SSc and MCTD. Clin Exp Immunol. 2020. https://doi.org/10.1111/cei.13566.
Uzrail AH, Assaf AM, Abdalla SS. Correlations of expression levels of a panel of genes (IRF5, STAT4, TNFSF4, MECP2, and TLR7) and cytokine levels (IL-2, IL-6, IL-10, IL-12, IFN-gamma, and TNF-alpha) with systemic lupus erythematosus outcomes in jordanian patients. Biomed Res Int. 2019;2019:1703842.
Niwa Y, Nishiyama C, Nakano N, et al. Opposite effects of PU.1 on mast cell stimulation. Biochem Biophys Res Commun. 2008;375(1):95–100.
Kanno Y, Levi BZ, Tamura T, et al. Immune cell-specific amplification of interferon signaling by the IRF-4/8-PU.1 complex. J Interferon Cytokine Res. 2005;25(12):770–9.
Lee YH, Bae SC. Association between circulating transforming growth factor-beta1 level and polymorphisms in systemic lupus erythematosus and rheumatoid arthritis: a meta-analysis. Cell Mol Biol (Noisy-le-grand). 2017;63(1):53–9.
Sugimoto A, Kawakami R, Mikami N. Transcription factors downstream of IL-4 and TGF-beta signals: analysis by quantitative PCR, Western Blot, and Flow Cytometry. Methods Mol Biol. 2017;1585:141–53.
Jovanovic M, Rooney MS, Mertins P, et al. Immunogenetics. Dynamic profiling of the protein life cycle in response to pathogens[J]. Science. 2015;347(6226):1259038.
Liu Y, Beyer A, Aebersold R. On the dependency of cellular protein levels on mRNA abundance [J]. Cell. 2016;165(3):535–50.
Acknowledgements
We thank all the volunteers who generously participated in this study.
Funding
This article was supported by the National Natural Science Foundation of China (No. 81871271), Anhui Key Research and Development Program (Grant Number 1804b06020354) and the Fundamental Research Funds for the Central Universities (Grant Number WK 9110000148).
Author information
Authors and Affiliations
Contributions
NX conceived and designed the study, NX and XF performed the experiments, and XL reviewed and edited the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no competing interests.
Consent for publication
All authors read and approved the manuscript.
Ethical approval
The experimental protocol was established, according to the ethical guidelines of the Helsinki Declaration and was approved by the Human Ethics Committee of Anhui Provincial Hospital. Written informed consent was obtained from individual participants.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Xiang, N., Fang, X., Sun, XG. et al. Expression profile of PU.1 in CD4+T cells from patients with systemic lupus erythematosus. Clin Exp Med 21, 621–632 (2021). https://doi.org/10.1007/s10238-021-00717-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10238-021-00717-9