Abstract
Background
Tubular injury plays a crucial role in the pathogenesis of diabetic nephropathy (DN). It is well known that many microRNAs (miRNAs) exert crucial effects on tubular injury. This study intends to explore the effect of miR-142-3p on the apoptosis and oxidative stress of high glucose (HG)-treated renal tubular epithelial cells (HK-2) and its underlying mechanism.
Materials and methods
HK-2 cells were exposed to HG to mimic cell injury. MTT assays and flow cytometry analyses were conducted to measure cell viability and cell apoptosis, respectively. RT-qPCR and western blot analyses were carried out to detect RNA and protein levels, respectively. The levels of oxidative stress markers were evaluated by ELISA. The binding between miR-142-3p and biorientation of chromosomes in cell division 1 (BOD1) was validated by a luciferase reporter assay.
Result
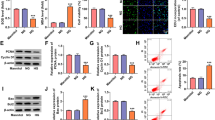
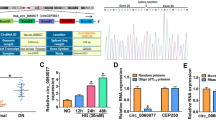
MiR-142-3p is low-expressed in HG-stimulated HK-2 cells. Functionally, miR-142-3p overexpression attenuates the apoptosis and oxidative stress of HG-stimulated HK-2 cells. Mechanistically, BOD1 was confirmed to be targeted by miR-142-3p in HK-2 cells. Moreover, BOD1 overexpression reversed the suppressive effect of miR-142-3p overexpression on the apoptosis and oxidative stress of HK-2 cells treated with HG.
Conclusion
MiR-142-3p ameliorates HG-induced renal tubular epithelial cell injury by targeting BOD1. The finding might provide novel insight into the role of miR-142-3p/BOD1 axis in DN treatment.





Similar content being viewed by others
References
Lin YC, Chang YH, Yang SY, Wu KD, Chu TS. Update of pathophysiology and management of diabetic kidney disease. J Formos Med Assoc. 2018;117(8):662–75. https://doi.org/10.1016/j.jfma.2018.02.007.
Qi C, Mao X, Zhang Z, Wu H. Classification and differential diagnosis of diabetic nephropathy. J Diabetes Res. 2017;2017:8637138. https://doi.org/10.1155/2017/8637138.
Papadopoulou-Marketou N, Chrousos GP, Kanaka-Gantenbein C. Diabetic nephropathy in type 1 diabetes: a review of early natural history, pathogenesis, and diagnosis. Diabetes Metab Res Rev. 2017. https://doi.org/10.1002/dmrr.2841.
Papadopoulou-Marketou N, Paschou SA, Marketos N, Adamidi S, Adamidis S, Kanaka-Gantenbein C. Diabetic nephropathy in type 1 diabetes. Minerva Med. 2018;109(3):218–28. https://doi.org/10.23736/s0026-4806.17.05496-9.
Ioannou K. Diabetic nephropathy: is it always there? Assumptions, weaknesses and pitfalls in the diagnosis. Hormones (Athens). 2017;16(4):351–61. https://doi.org/10.14310/horm.2002.1755.
Meza Letelier CE, San Martín Ojeda CA, Ruiz Provoste JJ, Frugone Zaror CJ. Pathophysiology of diabetic nephropathy: a literature review. Medwave. 2017;17(1):e6839. https://doi.org/10.5867/medwave.2017.01.6839.
Lu TX, Rothenberg ME. MicroRNA. J Allergy Clin Immunol. 2018;141(4):1202–7. https://doi.org/10.1016/j.jaci.2017.08.034.
Mohr AM, Mott JL. Overview of microRNA biology. Semin Liver Dis. 2015;35(1):3–11. https://doi.org/10.1055/s-0034-1397344.
Regazzi R. MicroRNAs as therapeutic targets for the treatment of diabetes mellitus and its complications. Expert Opin Ther Targets. 2018;22(2):153–60. https://doi.org/10.1080/14728222.2018.1420168.
Rosado JA, Diez-Bello R, Salido GM, Jardin I. Fine-tuning of microRNAs in type 2 diabetes mellitus. Curr Med Chem. 2019;26(22):4102–18. https://doi.org/10.2174/0929867325666171205163944.
Kumar M, Nath S, Prasad HK, Sharma GD, Li Y. MicroRNAs: a new ray of hope for diabetes mellitus. Protein Cell. 2012;3(10):726–38. https://doi.org/10.1007/s13238-012-2055-0.
Simonson B, Das S. MicroRNA therapeutics: the next magic bullet? Mini Rev Med Chem. 2015;15(6):467–74. https://doi.org/10.2174/1389557515666150324123208.
Yu J, Yu C, Feng B, Zhan X, Luo N, Yu X, et al. Intrarenal microRNA signature related to the fibrosis process in chronic kidney disease: identification and functional validation of key miRNAs. BMC Nephrol. 2019;20(1):336. https://doi.org/10.1186/s12882-019-1512-x.
Zanchi C, Macconi D, Trionfini P, Tomasoni S, Rottoli D, Locatelli M, et al. MicroRNA-184 is a downstream effector of albuminuria driving renal fibrosis in rats with diabetic nephropathy. Diabetologia. 2017;60(6):1114–25. https://doi.org/10.1007/s00125-017-4248-9.
Zhao Y, Yin Z, Li H, Fan J, Yang S, Chen C, et al. MiR-30c protects diabetic nephropathy by suppressing epithelial-to-mesenchymal transition in db/db mice. Aging Cell. 2017;16(2):387–400. https://doi.org/10.1111/acel.12563.
Sun J, Zhao F, Zhang W, Lv J, Lv J, Yin A. BMSCs and miR-124a ameliorated diabetic nephropathy via inhibiting notch signalling pathway. J Cell Mol Med. 2018;22(10):4840–55. https://doi.org/10.1111/jcmm.13747.
Zhou L, Xu DY, Sha WG, Shen L, Lu GY, Yin X, et al. High glucose induces renal tubular epithelial injury via Sirt1/NF-kappaB/microR-29/Keap1 signal pathway. J Transl Med. 2015;13:352. https://doi.org/10.1186/s12967-015-0710-y.
Kantharidis P, Hagiwara S, Brennan E, McClelland AD. Study of microRNA in diabetic nephropathy: isolation, quantification and biological function. Nephrology (Carlton). 2015;20(3):132–9. https://doi.org/10.1111/nep.12374.
Rudnicki M, Beckers A, Neuwirt H, Vandesompele J. RNA expression signatures and posttranscriptional regulation in diabetic nephropathy. Nephrol Dial Transplant. 2015;30(Suppl 4):35–42. https://doi.org/10.1093/ndt/gfv079.
Zeng Y, Feng Z, Liao Y, Yang M, Bai Y, He Z. Diminution of microRNA-98 alleviates renal fibrosis in diabetic nephropathy by elevating Nedd4L and inactivating TGF-β/Smad2/3 pathway. Cell Cycle. 2020;19(24):3406–18. https://doi.org/10.1080/15384101.2020.1838780.
Liu F, Zhang ZP, Xin GD, Guo LH, Jiang Q, Wang ZX. miR-192 prevents renal tubulointerstitial fibrosis in diabetic nephropathy by targeting Egr1. Eur Rev Med Pharmacol Sci. 2018;22(13):4252–60. https://doi.org/10.26355/eurrev_201807_15420.
Zhao D, Jia J, Shao H. miR-30e targets GLIPR-2 to modulate diabetic nephropathy: in vitro and in vivo experiments. J Mol Endocrinol. 2017;59(2):181–90. https://doi.org/10.1530/jme-17-0083.
Gholaminejad A, Abdul Tehrani H, Gholami FM. Identification of candidate microRNA biomarkers in renal fibrosis: a meta-analysis of profiling studies. Biomarkers. 2018;23(8):713–24. https://doi.org/10.1080/1354750x.2018.1488275.
Liang YZ, Li JJ, Xiao HB, He Y, Zhang L, Yan YX. Identification of stress-related microRNA biomarkers in type 2 diabetes mellitus: a systematic review and meta-analysis. J Diabetes. 2020;12(9):633–44. https://doi.org/10.1111/1753-0407.12643.
Zhu H, Leung SW. Identification of microRNA biomarkers in type 2 diabetes: a meta-analysis of controlled profiling studies. Diabetologia. 2015;58(5):900–11. https://doi.org/10.1007/s00125-015-3510-2.
Zhang T, Ji C, Shi R. miR-142-3p promotes pancreatic β cell survival through targeting FOXO1 in gestational diabetes mellitus. Int J Clin Exp Pathol. 2019;12(5):1529–38.
Zhou L, Xu DY, Sha WG, Shen L, Lu GY, Yin X. Long non-coding MIAT mediates high glucose-induced renal tubular epithelial injury. Biochem Biophys Res Commun. 2015;468(4):726–32. https://doi.org/10.1016/j.bbrc.2015.11.023.
Vejnar CE, Zdobnov EM. MiRmap: comprehensive prediction of microRNA target repression strength. Nucleic Acids Res. 2012;40(22):11673–83. https://doi.org/10.1093/nar/gks901.
Kertesz M, Iovino N, Unnerstall U, Gaul U, Segal E. The role of site accessibility in microRNA target recognition. Nat Genet. 2007;39(10):1278–84. https://doi.org/10.1038/ng2135.
Miranda KC, Huynh T, Tay Y, Ang YS, Tam WL, Thomson AM, et al. A pattern-based method for the identification of MicroRNA binding sites and their corresponding heteroduplexes. Cell. 2006;126(6):1203–17. https://doi.org/10.1016/j.cell.2006.07.031.
Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120(1):15–20. https://doi.org/10.1016/j.cell.2004.12.035.
Qi C, Mao X. Classification and Differential Diagnosis of Diabetic. Nephropathy. 2017;2017:8637138. https://doi.org/10.1155/2017/8637138.
Flyvbjerg A. The role of the complement system in diabetic nephropathy. Nat Rev Nephrol. 2017;13(5):311–8. https://doi.org/10.1038/nrneph.2017.31.
Tang J, Yao D, Yan H, Chen X, Wang L, Zhan H. The role of MicroRNAs in the pathogenesis of diabetic nephropathy. Int J Endocrinol. 2019;2019:8719060. https://doi.org/10.1155/2019/8719060.
Lu Y, Liu D, Feng Q, Liu Z. Diabetic nephropathy: perspective on extracellular vesicles. Front Immunol. 2020;11:943. https://doi.org/10.3389/fimmu.2020.00943.
Sagoo MK, Gnudi L. Diabetic nephropathy: is there a role for oxidative stress? Free Radical Biol Med. 2018;116:50–63. https://doi.org/10.1016/j.freeradbiomed.2017.12.040.
Magee C, Grieve DJ, Watson CJ, Brazil DP. Diabetic Nephropathy: a tangled web to unweave. Cardiovasc Drugs Ther. 2017;31(5–6):579–92. https://doi.org/10.1007/s10557-017-6755-9.
Jha JC, Banal C, Chow BS, Cooper ME, Jandeleit-Dahm K. Diabetes and kidney disease: role of oxidative stress. Antioxid Redox Signal. 2016;25(12):657–84. https://doi.org/10.1089/ars.2016.6664.
Bao L, Li J, Zha D, Zhang L, Gao P, Yao T, et al. Chlorogenic acid prevents diabetic nephropathy by inhibiting oxidative stress and inflammation through modulation of the Nrf2/HO-1 and NF-ĸB pathways. Int Immunopharmacol. 2018;54:245–53. https://doi.org/10.1016/j.intimp.2017.11.021.
Hou Y, Shi Y, Han B, Liu X, Qiao X, Qi Y, et al. The antioxidant peptide SS31 prevents oxidative stress, downregulates CD36 and improves renal function in diabetic nephropathy. Nephrol Dial Transplant. 2018;33(11):1908–18. https://doi.org/10.1093/ndt/gfy021.
Al-Rasheed NM, Al-Rasheed NM, Bassiouni YA, Hasan IH, Al-Amin MA, Al-Ajmi HN, et al. Simvastatin ameliorates diabetic nephropathy by attenuating oxidative stress and apoptosis in a rat model of streptozotocin-induced type 1 diabetes. Biomed Pharmacother. 2018;105:290–8. https://doi.org/10.1016/j.biopha.2018.05.130.
Wang Y, Li Y, Yang Z, Wang Z, Chang J, Zhang T, et al. Pyridoxamine treatment of HK-2 human proximal tubular epithelial cells reduces oxidative stress and the inhibition of autophagy induced by high glucose levels. Med Sci Monit. 2019;25:1480–8. https://doi.org/10.12659/msm.914799.
Chen F, Sun Z, Zhu X, Ma Y. Astilbin inhibits high glucose-induced autophagy and apoptosis through the PI3K/Akt pathway in human proximal tubular epithelial cells. Biomed Pharmacother. 2018;106:1175–81. https://doi.org/10.1016/j.biopha.2018.07.072.
Kimura T, Takabatake Y, Takahashi A, Kaimori JY, Matsui I, Namba T, et al. Autophagy protects the proximal tubule from degeneration and acute ischemic injury. J Am Soc Nephrol. 2011;22(5):902–13. https://doi.org/10.1681/asn.2010070705.
Kitada M, Ogura Y, Monno I, Koya D. Regulating autophagy as a therapeutic target for diabetic nephropathy. Curr Diab Rep. 2017;17(7):53. https://doi.org/10.1007/s11892-017-0879-y.
Wei Z, Qin X, Kang X, Zhou H, Wang S, Wei D. MiR-142-3p inhibits adipogenic differentiation and autophagy in obesity through targeting KLF9. Mol Cell Endocrinol. 2020;518: 111028. https://doi.org/10.1016/j.mce.2020.111028.
Su Q, Liu Y, Lv XW, Ye ZL, Sun YH, Kong BH, et al. Inhibition of lncRNA TUG1 upregulates miR-142-3p to ameliorate myocardial injury during ischemia and reperfusion via targeting HMGB1- and Rac1-induced autophagy. J Mol Cell Cardiol. 2019;133:12–25. https://doi.org/10.1016/j.yjmcc.2019.05.021.
Plotnikova OM, Skoblov MY. Efficiency of the miRNA- mRNA interaction prediction programs. Mol Biol. 2018;52(3):543–54. https://doi.org/10.7868/s0026898418030187.
Yang Z, Guo Z, Dong J, Sheng S, Wang Y, Yu L, et al. miR-374a regulates inflammatory response in diabetic nephropathy by targeting MCP-1 expression. Front Pharmacol. 2018;9:900. https://doi.org/10.3389/fphar.2018.00900.
Wang J, Duan L, Guo T, Gao Y, Tian L, Liu J, et al. Downregulation of miR-30c promotes renal fibrosis by target CTGF in diabetic nephropathy. J Diabetes Complicat. 2016;30(3):406–14. https://doi.org/10.1016/j.jdiacomp.2015.12.011.
Qin B, Shu Y, Long L, Li H, Men X, Feng L, et al. MicroRNA-142-3p induces atherosclerosis-associated endothelial cell apoptosis by directly targeting rictor. Cell Physiol Biochem. 2018;47(4):1589–603. https://doi.org/10.1159/000490932.
Sukma Dewi I, Celik S, Karlsson A, Hollander Z, Lam K, McManus JW, et al. Exosomal miR-142-3p is increased during cardiac allograft rejection and augments vascular permeability through down-regulation of endothelial RAB11FIP2 expression. Cardiovasc Res. 2017;113(5):440–52. https://doi.org/10.1093/cvr/cvw244.
Pan D, Du Y, Ren Z, Chen Y, Li X, Wang J, et al. Radiation induces premature chromatid separation via the miR-142-3p/Bod1 pathway in carcinoma cells. Oncotarget. 2016;7(37):60432–45. https://doi.org/10.18632/oncotarget.11080.
Di J, Rutherford S, Chu C. Review of the cervical cancer burden and population-based cervical cancer screening in China. Asian Pac J Cancer Prev. 2015;16(17):7401–7. https://doi.org/10.7314/apjcp.2015.16.17.7401.
Porter IM, Schleicher K, Porter M, Swedlow JR. Bod1 regulates protein phosphatase 2A at mitotic kinetochores. Nat Commun. 2013;4:2677. https://doi.org/10.1038/ncomms3677.
Gao Y, Zhao H, Li Y. LncRNA MCM3AP-AS1 regulates miR-142-3p/HMGB1 to promote LPS-induced chondrocyte apoptosis. BMC Musculoskelet Disord. 2019;20(1):605. https://doi.org/10.1186/s12891-019-2967-4.
Funding
This research was supported by Henan Science and Technology Research Project (No. 162102310287), Henan health and Family Planning Commission Provincial Ministry Co Construction Project (No.201701022), Henan Medical Science and Technology Research Project (No.2018020395).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors report no declarations of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
About this article
Cite this article
Zhao, N., Luo, Q., Lin, R. et al. MiR-142-3p ameliorates high glucose-induced renal tubular epithelial cell injury by targeting BOD1. Clin Exp Nephrol 25, 1182–1192 (2021). https://doi.org/10.1007/s10157-021-02102-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10157-021-02102-y